Figure 5.
The Role of Condensins in Chromosome Segregation Is Not Limited to Resolution of Sister Chromatids
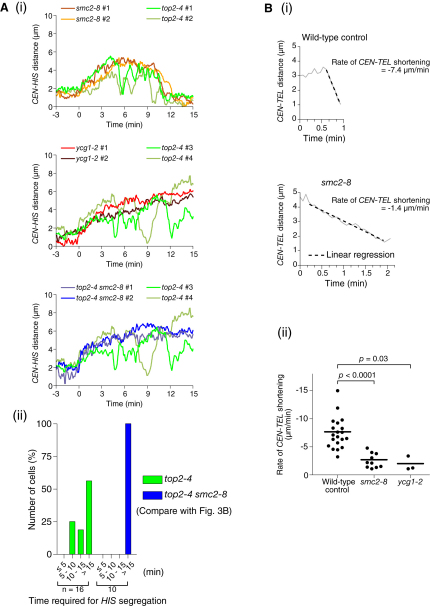
(A) Condensin mutants show different behaviors of a chromosome arm locus, compared with a top2 mutant. smc2-8 (T3829), ycg1-2 (T3992), top2-4 (T3794), and top2-4 smc2-8 (T3936) cells with Pgal-CDC20 TetR-GFP tetOs (integrated at three loci as in Figure 1A) were treated and analyzed as in Figure 3. (Ai) Graphs show the CEN-HIS distances (time 0: CEN-CEN distance [not shown] reached 3 μm), for two representative top2-4 cells (#1 and 2) that completed HIS segregation with similar timing to the majority of smc2-8 cells (top); and for two representative top2-4 cells (#3 and 4) that did not complete HIS segregation, similarly to many ycg1-2 cells (middle). Finally, two top2-4 cells (#3 and 4) were compared with two representative top2-4 smc2-8 cells (bottom). The smc2-8 #1 and ycg1-2 #1 cells were also analyzed in Figure 3A. Figures S3B and S5A and Movies S2–S4 and S7 also concern the cells shown here. See Figure 3A (blue line) for the change in CEN-HIS distance in a representative “wild-type” cell. (Aii) The percentage of cells, in which HIS segregation completed (CEN-HIS distance became <1.8 μm without subsequently exceeding 3 μm) within the indicated time window. Compare with the results in “wild-type,” smc2-8, and ycg1-2 cells, shown in Figure 3B. n = number of observed cells.
(B) The speed of telomere segregation is lower in condensin mutants. smc2-8 (T3829), ycg1-2 (T3992), and control wild-type cells (T3790; see their genotypes in A and Figure 3) were treated and analyzed as in Figure 3. (Bi) Changes in CEN-TEL distance in representative cells. Time 0 is set arbitrarily. (Bii) The speed of TEL segregation (shortening of CEN-TEL distance) toward the bud in individual cells. Thick lines indicate mean values.