Figure 6.
Condensins Play Active Roles in Chromosome Recoiling Independently of Sister Chromatid Resolution/Separation
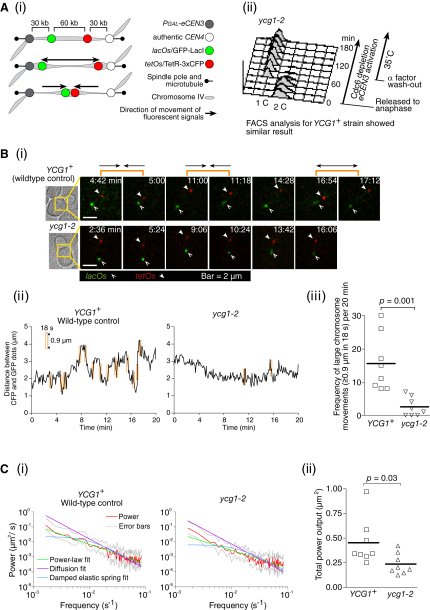
(A) Making an unreplicated chromosome with two centromeres. YCG1+ (condensin wild-type, T5822) and ycg1-2 (T5823) cells with Pgal-CDC6 Pmet-CDC20 Pgal-eCEN3 (ectopic CEN3; integrated 120 kb to the left of the authentic CEN4) TetR-3×CFP GFP-LacI lacOs tetOs (integrated as shown in [Ai]) were arrested in metaphase by Cdc20 depletion and then released to anaphase (0 min in [Aii] FACS analyses) by re-expression of Cdc20 at 25°C. From 30 min before re-expression of Cdc20, Cdc6 expression was inhibited and eCEN3 was activated in glucose-containing medium. Cells were then arrested by α factor treatment, followed by release (90 min in [Aii]) into fresh medium at 35°C. After 80 min, GFP and CFP images were acquired every 6 s for 20 min.
(B) An unreplicated dicentric chromosome shows condensin-dependent stretching and recoiling. (Bi) Representative time-lapse images (time 0, start of image acquisition) showing tetOs (CFP) in red and lacOs (GFP) in green. Orange brackets indicate examples of stretching and recoiling of the region between the CFP and GFP dots. (Bii) Changes in the distance between the two dots in the cells shown in (Bi). (Biii) Frequency of large changes (≥0.9 μm within 18 s) in the distance between the two dots in individual cells. Thick lines indicate mean values.
(C) Power spectra analyses for the oscillation of the CFP and GFP dots. (Ci) Using discrete Fourier transforms, power in the oscillation (red) was plotted as function of frequency of oscillation. Error bars (gray) show 90% confidence of the power. The power was fitted by curves, based on a motion of power-law (green), diffusion (purple), or damped elastic spring (cyan). See more detail in Supplemental Note for Power Spectrum Analyses. (Cii) Total power (variance) in individual cells. Thick lines indicate mean values.