Abstract
The most abundant chemical modification on RNA is isomerization of uridine, or pseudouridylation, catalyzed by pseudouridine synthases. The catalytic mechanism of this essential process remains largely speculative, partly due to the lack of knowledge of the pre-reactive state that is important to identification of reactive chemical moieties. In the present study, we showed by the orthogonal space random walk free energy simulation method that the pre-reactive states of uridine and its reactive derivative, 5-fluorouridine, bound to a ribonucleoprotein particle (RNP) pseuoduridine synthase strongly prefer the syn glycosidic bond conformation while that of the nonreactive 5-bromouridine-containing substrate is largely populated in the anti conformational state. A high resolution crystal structure of the 5-bromouridine containing substrate bound to the RNP pseuoduridine synthase and enzyme activity assay confirmed the anti nonreactive conformation and provides molecular basis for its confinement. The observed preference for the syn pre-reactive state by the enzyme-bound uridine may help to distinguish among currently proposed mechanisms.
Despite its prevalence1,2 and demonstrated importance in RNA structure3-5 and function6,7, the catalytic mechanism of pseudouridylation remains largely speculative, partly due to the lack of knowledge of the pre-reactive state critical to identification of reactive chemical moieties. This process is thought to begin with the cleavage of the N-glycosyl bond (ring cleavage), followed by a 180° rotation of the uracil base while still enzyme-bound (ring rotation), then reattachment of the ring at C5 (ring reattachment) and finally deprotonation of C58-10 (Fig. 1). Cofactors are not known to be required for any of these steps.
Figure 1.
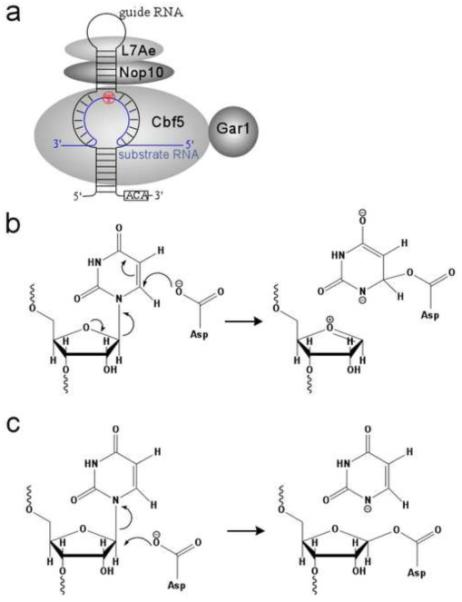
Schematics of the two proposed mechanisms for pseudouridylation and enzyme activity assay results. (a) Michael addition mechanism. (b) Acylal mechanism. The two mechanisms share the same catalytic steps and the catalytic Asp residue but differ in the role Asp plays in ring cleavage.
In bacteria, approximately a dozen uridine residues in transfer RNA (tRNA) and ribosomal RNA (rRNA) are modified by six families of pseudouridine synthases: TruA, TruB, TruD, RluA, RsuA, and Pus1011-19. Each of these pseudouridine synthases is responsible for modifying one or several specific uridine nucleotide(s) in tRNA or rRNA. In Archaea and Eukarya where rRNA and small nuclear RNA (snRNA) are extensively modified, pseudouridylation is largely carried out by the box H/ACA ribonucleoprotein particle (RNP) pseudouridine synthases20-24. Unlike the stand-alone pseudouridine synthases, box H/ACA RNP pseudouridine synthases are multi-subunit enzymes and are comprised of four protein and one RNA subunits (Figure 1a). The four protein subunits include Cbf5, Nop10, Gar1, and Nhp2 (L7Ae in archaea). Cbf5 shares sequence motifs and a structural similarity with the TruB family of pseudouridine synthases and is the catalytic subunit of the RNP enzyme. The RNA subunit belongs to the class of box H/ACA noncoding RNA and is characterized by the hairpin-like secondary structure and the strictly conserved ACA trinucleotide at its 3’ end20-24. The substrate RNA is captured by the RNP enzyme through its base pairing with the central internal loop (pseudouridine pocket) of the box H/ACA RNA (Figure 1a).
Regardless of their substrate specificity and enzyme composition, however, all families of pseudouridine synthases contain a well-conserved catalytic domain and a catalytic aspartate residue9,25,26. Mutation of catalytic Asp to Asn in bacterial TruB and TruA pseudouridine synthases resulted in complete loss of enzyme activity9,27. Furthermore, mutation of Asp to Ala in an archaeal H/ACA RNP also abolished the modification activity28, suggesting that this residue is essential to catalysis.
Two mechanisms that invoke Asp as a nucleophile have been proposed for the catalytic process. In the Michael addition mechanism (Fig. 1b), Asp attacks the RNA ring atom C6, leading to an Asp-pyrimidine covalent adduct. The strongest evidence supporting this mechanism is the observation that on a RNA substrate containing 5’-fluorouridine (5FU), E. coli TruA and RluA either cross-link to or strongly interact with the hydrated product, 5-fluoro-6-hydroxpseudouridine (5FhΨ)29,30. In opposition to the proposed Michael addition mechanisms, however, crystal structures of TruB, RluA, and the H/ACA pseudouridine synthase bound to 5FU substituted RNA substrates do not show a covalent intermediate, although it was argued in the case of the RluA-RNA complex that X-ray radiation used for diffraction studies dissolved the possible covalent linkage17,19,31,32. Furthermore, the bound 5FhΨ in all cases has its C6 pointing away from the γ-carboxyl group of the catalytic Asp. The unfavorable ring orientation is also observed in the complex of the Asp-to-Asn TruB mutant bound to a 5FU-containing RNA substrate33. The second proposed mechanism is the acylal mechanism (Fig. 1c) in which the catalytic Asp attacks the sugar atom C1’ to form the acylal intermediate that stabilizes the oxocarbonium ion. This mechanism should be less sensitive to the ring orientation than to the distance between Asp and C1’. Elucidation of the pseuoduiridine synthase mechanism thus requires an assessment of the glycosidic bond conformation of uridine in its pre-reactive state.
Previously, we obtained a cocrystal structure of a functional H/ACA RNP pseudouridine synthase bound to a guide RNA and a 5FU-containing substrate RNA32. This structure made possible for us to explore the theoretical pre-reactive state of the substrate. We carried out free energy simulation on the box H/ACA RNP complex containing the wild-type and 5FU substrates using the previously developed Orthogonal Space Random Walk (OSRW) algorithm34,35. As detailed in the supplementary information, we utilized a novel “alchemical” transition36,37 scheme to realize the syn to anti conformational transformation about the glycosidic bond of uridine (Fig. 2a). The previously determined structure containing 5FhΨ was used to build the starting structures for the simulation by substituting 5FhΨ for uridine base. We found that for both the wild-type and 5FU-subsituted substrates, the syn conformation is preferred over anti by −3.5 kcal/mol and −2.5 kcal/mol, respectively. This is in contrast to the anti conformation of 5-fluorouridine captured in TruB D48N crystal structure containing 5FU33, suggesting that the D48N mutation may sufficiently disturb the micro environment of active site to favor the anti conformation.
Figure 2.
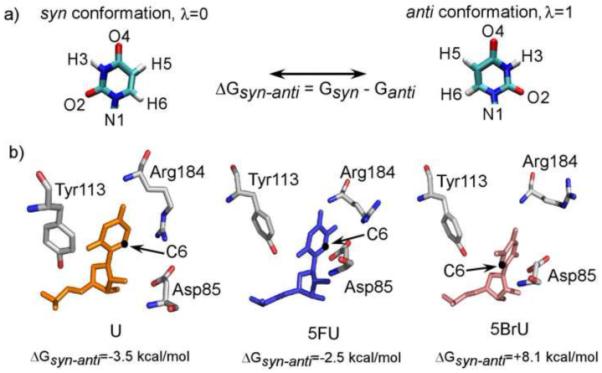
Computationally identified low energy structures of uridine (orange), 5FhΨ (blue) and 5BrU (pink). The syn and anti conformations and their free energy difference are defined as shown in a) and are represented by a “coupling parameter”, for the purpose of calculation of free energy difference. Computed difference free energy after convergence is shown below each complex in b). Details of free energy simulation are included in the supplementary material.
The syn conformation places the ring C6 atom close to the catalytic Asp85 (Fig. 2b). In order to test the importance of the syn conformation to catalysis, we carried out the same free energy simulation on 5-bromouridine-containing substrate (5BrU) bound to the H/ACA RNP. Bromine is less electronegative but bulkier in size than fluorine. Consistently, 5BrU was found to prefer the anti over the syn conformation by more than 8 kcal/mol (Fig. 2b), suggesting it may be defective in the ring cleavage step.
To determine if 5BrU is a substrate for the pseudouridine synthase, we used the DNA splint technique for constructing both wild-type and 5BrU-containing substrate RNA from two synthetic oligos38 with a 32P-label on the 5’ position of the target uridine or modified uridine. The integrity of the ligated substrate RNA was checked on a denaturing polyacrylamide gel (data not shown). After pseudouridylation assays with saturating amount of H/ACA RNP pseudouridine synthase and nuclease P1 digestion of RNA substrates, formation of uridine (or modified uridine) isomers was detected by thin layer chromatography. As shown in Figure 3a, 5BrU completely inhibited isomerization.
Figure 3.
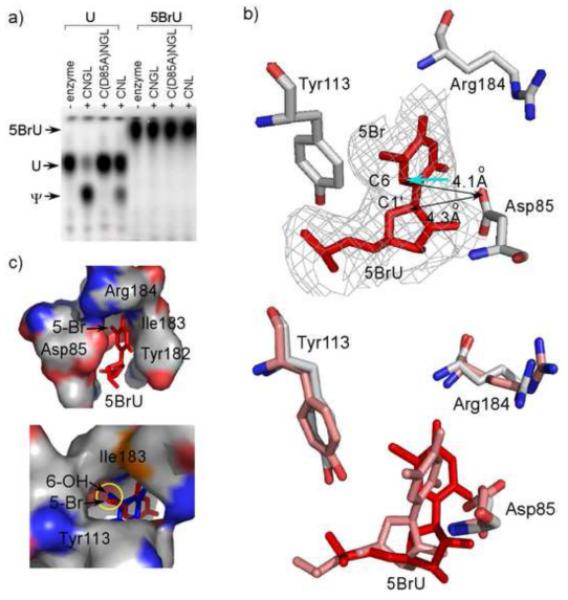
Enzyme activity assay (a) and structural study results (b-c) of 5BrU bound to H/ACA ribonucleoprotein particle (RNP) pseudouridine synthase. a). Thin layer chromatography separation of reacted and digested wild-type (U) or 5BrU substrate nucleotides. “C” denotes Cbf5, “N” denotes Nop10, “G” denotes “Gar1”, and “L” denotes L7Ae. D85A is the catalytically deficient mutant of Cbf5. b). (upper) Crystal structure of the active site of the 5BrU substrate bound to H/ACA RNP. Omitted 3Fo-2Fc map is shown at 1.0σ around the target nucleotide. (lower) Comparsion of the computated low energy structure (pink) to the crystal structure (red). c). (upper) 5BrU (red) is tightly bound by the active site residues (surface). (lower) Superimposed 5FhΨ (blue) shows a similar location of the 6-hydroxyl group as 5-bomide. Experimental details for enzyme activity assay, protein crystallization and structure determination are included in the supplementary material.
To further provide evidence for the reactivity of 5BrU with the enzyme, we obtained a crystal structure of 5BrU in complex with the Pyrococcus furiosus H/ACA RNP that contains Cbf5, Nop10, L7Ae and a guide RNA at 2.9 Å. The crystallographic data are listed in table 1 of supplementary material. Its global structure bear strong similarity to the previously determined 5FhΨ-bound RNP structure31,32. However, sigmaA-weighted electron density maps of 5BrU clearly revealed that it did not react to the enzyme and is in the computationally predicted anti conformation (Fig. 3b). Although its C1’ atom has a similar distance to the carbonyl group of the catalytic Asp85 (4.3 Å) as that of 5FhΨ, its C6 atom is pointed away from Asp85. Even if Asp85 could facilitate a nucleophilic attack on C6 in this orientation (shown as the cyan arrow in Fig. 3b), it would result in the minor trans 6-hydroxyl stereoisomer39. Therefore, 5BrU is confined to an orientation that does not permit the initial attack to take place. The bromide atom is primarily responsible for the change as in its absence the anti conformation is preferred. The bromide atom is in close contact with several polar and aliphatic groups of the active site. These include the hydroxyl group of Tyr113, the amide of Ile183 and the aliphatic groups of Ile183 (Fig. 3c). Strikingly, the O6 atom in the bound 5FhΨ is located in exactly the same site and establishes similar extensive interactions with the enzyme, which likely serves to hyperstabilize the reaction intermediate (Fig. 3c).
We have demonstrated a clear preference for the syn conformation by uridine in the microenvironement provided by the H/ACA RNP pseudouridine synthase. Perturbation of this conformation resulted in inhibition of the activity. These results highlight an important role of the pre-reactive glycosidic bond conformation in catalysis. Significantly, the predicted pre-reactive state conformation suggests a preference for a C6-based catalytic mechanism. Regardless the actual mechanism of nucleophilic attack, C6-based schemes are consistent with formation of the 5FhΨ intermediate8,29 and the acquired 6-hydroxyl group from the aqueous solution8.
Supplementary Material
ACKNOWLEDGMENT
This work was supported in part by a National Institutes of Health grant R01 GM66958-01 (H.L), a National Science Foundation grant 0919983 (W.Y.) and two American Heart Association, Florida/Puerto Rico Affiliate, predoctoral fellowships (0815267E to J. Z and 0815267E to B.L., respectively). X-ray diffraction data were collected from the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Footnotes
ACCESSION NUMBERS:
Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 3LWO.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–51. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 2.Grosjean H, Benne R. Modification and editing of RNA. ASM Press; Washington, DC: 1998. [Google Scholar]
- 3.Arnez JG, Steitz TA. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry. 1994;33:7560–7. doi: 10.1021/bi00190a008. [DOI] [PubMed] [Google Scholar]
- 4.Newby MI, Greenbaum NL. Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc. Natl. Acad. Sci. USA. 2002;99:12697–702. doi: 10.1073/pnas.202477199. Epub 2002 Sep 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–6. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. Embo J. 1998;17:5783–95. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell. 2003;11:425–35. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- 8.Spedaliere CJ, Ginter JM, Johnston MV, Mueller EG. The pseudouridine synthases: revisiting a mechanism that seemed settled. J. Am. Chem. Soc. 2004;126:12758–9. doi: 10.1021/ja046375s. [DOI] [PubMed] [Google Scholar]
- 9.Huang L, Pookanjanatavip M, Gu X, Santi DV. A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry. 1998;37:344–51. doi: 10.1021/bi971874+. [DOI] [PubMed] [Google Scholar]
- 10.Zhao X, Horne DA. The role of cysteine residues in the rearrangement of uridine to pseudouridine catalyzed by pseudouridine synthase I. J. Biol. Chem. 1997;272:1950–5. doi: 10.1074/jbc.272.3.1950. [DOI] [PubMed] [Google Scholar]
- 11.Sivaraman J, Iannuzzi P, Cygler M, Matte A. Crystal structure of the RluD pseudouridine synthase catalytic module, an enzyme that modifies 23S rRNA and is essential for normal cell growth of Escherichia coli. J. Mol. Biol. 2004;335:87–101. doi: 10.1016/j.jmb.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Kaya Y, Del Campo M, Ofengand J, Malhotra A. Crystal structure of TruD, a novel pseudouridine synthase with a new protein fold. J. Biol. Chem. 2004;279:18107–10. doi: 10.1074/jbc.C400072200. [DOI] [PubMed] [Google Scholar]
- 13.Hoang C, Ferre-D’Amare AR. Crystal structure of the highly divergent pseudouridine synthase TruD reveals a circular permutation of a conserved fold. RNA. 2004;10:1026–33. doi: 10.1261/rna.7240504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sivaraman J, Sauve V, Larocque R, Stura EA, Schrag JD, Cygler M, Matte A. Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP. Nat. Struct. Biol. 2002;9:353–8. doi: 10.1038/nsb788. [DOI] [PubMed] [Google Scholar]
- 15.Dong X, Bessho Y, Shibata R, Nishimoto M, Shirouzu M, Kuramitsu S, Yokoyama S. Crystal structure of tRNA pseudouridine synthase TruA from Thermus thermophilus HB8. RNA Biol. 2006;3:115–22. doi: 10.4161/rna.3.3.3286. [DOI] [PubMed] [Google Scholar]
- 16.Del Campo M, Ofengand J, Malhotra A. Crystal structure of the catalytic domain of RluD, the only rRNA pseudouridine synthase required for normal growth of Escherichia coli. RNA. 2004;10:231–9. doi: 10.1261/rna.5187404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoang C, Chen J, Vizthum CA, Kandel JM, Hamilton CS, Mueller EG, Ferre-D’Amare AR. Crystal structure of pseudouridine synthase RluA: indirect sequence readout through protein-induced RNA structure. Mol. Cell. 2006;24:535–45. doi: 10.1016/j.molcel.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 18.McCleverty CJ, Hornsby M, Spraggon G, Kreusch A. Crystal structure of human Pus10, a novel pseudouridine synthase. J. Mol. Biol. 2007;373:1243–54. doi: 10.1016/j.jmb.2007.08.053. [DOI] [PubMed] [Google Scholar]
- 19.Hoang C, Ferre-D’Amare AR. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell. 2001;107:929–39. doi: 10.1016/s0092-8674(01)00618-3. [DOI] [PubMed] [Google Scholar]
- 20.Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–34. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 21.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 22.Decatur WA, Fournier MJ. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 2003;278:695–8. doi: 10.1074/jbc.R200023200. [DOI] [PubMed] [Google Scholar]
- 23.Baker DL, Youssef OA, Chastkofsky MI, Dy DA, Terns RM, Terns MP. RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes Dev. 2005;19:1238–48. doi: 10.1101/gad.1309605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meier UT. The many facets of H/ACA ribonucleoproteins. Chromosoma. 2005;114:1–14. doi: 10.1007/s00412-005-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamma T, Ferre-D’Amare AR. Pseudouridine synthases. Chem. Biol. 2006;13:1125–35. doi: 10.1016/j.chembiol.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 26.Koonin EV. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res. 1996;24:2411–5. doi: 10.1093/nar/24.12.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramamurthy V, Swann SL, Paulson JL, Spedaliere CJ, Mueller EG. Critical aspartic acid residues in pseudouridine synthases. J. Biol. Chem. 1999;274:22225–30. doi: 10.1074/jbc.274.32.22225. [DOI] [PubMed] [Google Scholar]
- 28.Charpentier B, Muller S, Branlant C. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res. 2005;33:3133–44. doi: 10.1093/nar/gki630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu X, Liu Y, Santi DV. The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluorouracil-tRNA. Proc. Natl. Acad. Sci. USA. 1999;96:14270–5. doi: 10.1073/pnas.96.25.14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamilton CS, Greco TM, Vizthum CA, Ginter JM, Johnston MV, Mueller EG. Mechanistic investigations of the pseudouridine synthase RluA using RNA containing 5-fluorouridine. Biochemistry. 2006;45:12029–38. doi: 10.1021/bi061293x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duan J, Li L, Lu J, Wang W, Ye K. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol. Cell. 2009;34:427–39. doi: 10.1016/j.molcel.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Liang B, Zhou J, Kahen E, Terns RM, Terns MP, Li H. Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA. Nat. Struct. Mol. Biol. 2009;16:740–6. doi: 10.1038/nsmb.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoang C, Hamilton CS, Mueller EG, Ferre-D’Amare AR. Precursor complex structure of pseudouridine synthase TruB suggests coupling of active site perturbations to an RNA-sequestering peripheral protein domain. Protein Sci. 2005;14:2201–6. doi: 10.1110/ps.051493605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng LQ, Chen MG, Yang W. Simultaneous escaping of explicit and hidden free energy barriers: Application of the orthogonal space random walk strategy in generalized ensemble based conformational sampling. J. Chem. Phys. 2009;130 doi: 10.1063/1.3153841. [DOI] [PubMed] [Google Scholar]
- 35.Zheng LQ, Chen MG, Yang W. Random walk in orthogonal space to achieve efficient free-energy simulation of complex systems. Proc. Natl. Acad. Sci. USA. 2008;105:20227–20232. doi: 10.1073/pnas.0810631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tembe BL, Mccammon JA. Ligand Receptor Interactions. Computers & Chemistry. 1984;8:281–283. [Google Scholar]
- 37.Kollman P. Free-Energy Calculations - Applications to Chemical and Biochemical Phenomena. Chemical Reviews. 1993;93:2395–2417. [Google Scholar]
- 38.Moore MJ, Sharp PA. Site-specific modification of pre-mRNA: the 2′-hydroxyl groups at the splice sites. Science. 1992;256:992–7. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- 39.Spedaliere CJ, Mueller EG. Not all pseudouridine synthases are potently inhibited by RNA containing 5-fluorouridine. RNA. 2004;10:192–9. doi: 10.1261/rna.5100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


