Abstract
Background:
The APOE ε4 allele is an established risk factor for Alzheimer disease (AD), yet findings are mixed for how early its effects are manifest. One reason for the mixed results could be the presence of interaction effects with other AD risk factors. Increasing evidence indicates that testosterone may play a significant role in the development of AD. The aim of the present study was to examine the potential interaction of testosterone and APOE genotype with respect to hippocampal volume in middle age.
Methods:
Participants were men from the Vietnam Era Twin Study of Aging (n = 375). The mean age was 55.9 years (range 51–59). Between-group comparisons were performed utilizing a hierarchical linear mixed model that adjusted for the nonindependence of twin data.
Results:
A significant interaction was observed between testosterone and APOE genotype (ε4-negative vs ε4-positive). Those with both low testosterone (≥1 SD below the mean) and an ε4-positive status had the smallest hippocampal volumes, although comparisons with normal testosterone groups were not significant. However, individuals with low testosterone and ε4-negative status had significantly larger hippocampal volumes relative to all other groups. A main effect of APOE genotype on hippocampal volume was observed, but only when the APOE-by-testosterone interaction was present.
Conclusions:
These findings demonstrate an interaction effect between testosterone and the APOE ε4 allele on hippocampal volume in middle-aged men, and they may suggest 2 low testosterone subgroups. Furthermore, these results allude to potential gene–gene interactions between APOE and either androgen receptor polymorphisms or genes associated with testosterone production.
GLOSSARY
- AD
= Alzheimer disease;
- AR
= androgen receptor;
- BMI
= body mass index;
- CI
= confidence interval;
- ICV
= intracranial volume;
- MGH
= Massachusetts General Hospital;
- UCSD
= University of California, San Diego;
- VET
= Vietnam Era Twin;
- VETSA
= Vietnam Era Twin Study of Aging.
The APOE ε4 allele is a major risk factor for late-onset Alzheimer disease (AD),1 but evidence for its association in middle-aged samples with brain structures, especially the hippocampus, and cognitive functioning has been mixed.2,3 One reason for mixed results in genetic studies may be that interactions with other biologic or environmental factors have obscured main effects.4 Testosterone is one such biologic factor that may interact with APOE genotype.
In men, testosterone levels begin to decline as early as the 4th decade of life, and lower levels are predictive of AD and mild cognitive impairment.5,6 Animal studies have demonstrated that in the brain, especially the hippocampus, testosterone influences the production and deposition of β-amyloid, a key component of the neuronal plaques associated with AD,7 and in older men testosterone is positively associated with hippocampal blood flow.8 The binding of testosterone to androgen receptors (ARs) has been found to be markedly reduced in mice that express the ε4 allele,9 leading to the hypothesis that APOE genotype differentially effects regulation of the AR.10 Indeed, having an ε4 allele combined with low levels of testosterone has been found to be a significant predictor of AD.11
The aim of the present study was to test for interactive effects of APOE genotype and testosterone on hippocampal volume in middle-aged men. Along with being a primary site of AD neuropathology,12 the hippocampus is rich in ARs,13 making it vulnerable to the age-related decline in testosterone levels. Extrapolating from previous animal models,9,10 we hypothesized that an interaction between APOE genotype and testosterone would be present in humans such that being both ε4-positive and having low testosterone levels would be associated with smaller hippocampal volumes relative to other combinations of APOE genotype and testosterone level.
METHODS
Participants.
Data were obtained from participants in the Vietnam Era Twin Study of Aging (VETSA), a longitudinal study of cognitive and brain aging with a baseline in midlife.14 VETSA participants were randomly sampled from the Vietnam Era Twin (VET) Registry, a nationally distributed sample of male–male twin pairs who served in the United States military sometime between 1965 and 1975. The VET Registry's composition and method of ascertainment have been described elsewhere.15 In total, 1,237 men ages 51 to 60 participated in the VETSA project between 2003 and 2007. In comparison to census data, VETSA participants are similar in demographic and health characteristics to American men in their age range.16 Neuroimaging (n = 474) and endocrine (n = 783) data were collected concurrently between 2005 and 2007. The present analyses are based on data from 375 VETSA participants for whom neuroimaging, endocrine, and APOE genotyping data were available.
As part of the primary VETSA project, participants traveled to either the University of California, San Diego (UCSD) or Boston University for a day-long series of assessments. To be eligible for the primary VETSA project, both members of a twin pair had to agree to participate and be between the ages of 51 and 59 years at the time of recruitment. Aside from standard exclusion criterion for MRI studies (e.g., metal in the body), there were no additional eligibility requirements.
Standard protocol approvals, registrations, and patient consents.
Informed consent was obtained from all participants prior to data collection, and institutional review board approval was obtained at all participating institutions.
Procedures.
Testosterone collection and assay.
Testosterone was obtained via saliva collection on the assessment day, as well as on 2 days during a participant's typical week. These at-home samples were collected approximately 2 weeks prior to the assessment day. Samples were collected at waking, 30 minutes after waking, 10:00 am, 3:00 pm, and bedtime on all days. Project staff worked with the participants to individualize collection times to work schedules and wake-up times when necessary. Participants were mailed a saliva collection kit which included individualized instructions, labeled 4.5-mL Cryotube vials, Trident original sugarless gum, straws, tissues, a daily log, pen, reminder watch, and a storage container with an electronic track cap for detecting compliance with the protocol. Samples were sent via overnight mail to the University of California, Davis, for assay.
Samples were centrifuged at 3,000 rpm for 20 minutes to separate the aqueous component from mucins and other suspended particles. Salivary concentrations of free testosterone were estimated in duplicate using commercial radioimmunoassay kits (Beckman Coulter Inc., formerly Diagnostics Systems Laboratories, Webster, TX). Assay procedures were identical to those outlined by Granger and colleagues.17 Intraassay and interassay coefficients of variation were 3.141 pg/mL and 4.878 pg/mL. The least detectable dose for the assay was 1.3697 pg/mL. All samples from each participant were analyzed in the same assay; 1 to 3 individuals were included in the same assay batch. Assays were always performed without knowledge of the zygosity of the twin pair. Values greater than 3 SD above the mean waking testosterone level, the highest level of the day, were recoded as missing. Data from participants who reported taking testosterone supplements were also set to missing.
MRI acquisition and processing.
Acquisition parameters and postprocessing details are described in detail elsewhere.17 Briefly, neuroimaging was performed within 24 hours of the assessment day at either the UCSD Medical Center or Massachusetts General Hospital (MGH). Images were acquired on Siemens 1.5-T scanners. Although scanners were not identical, scanning sequences were designed for use across scanners and vendors. Sagittal T1-weighted magnetization-prepared rapid gradient echo sequences were employed with inversion time = 1,000 msec, echo time = 3.31 msec, repetition time = 2,730 msec, flip angle = 7°, slice thickness = 1.33 mm, voxel size 1.3 × 1.0 × 1.3 mm. Raw DICOM MRI scans from both sites were downloaded to MGH for postprocessing and quality control.
Hippocampal volumes were obtained using segmentation methods based on the publically available FreeSurfer software package.18 The automated, fully 3-dimensional whole brain segmentation procedure uses a probabilistic atlas and applies a Bayesian classification rule to assign a neuroanatomic label to each voxel. This process required only qualitative review to ensure no technical failure of the application. We created a VETSA-specific atlas, and automated volumetric measurements based on this atlas were within the 99% confidence interval (CI) with respect to the gold standard manual measurements.19 Direct comparisons of FreeSurfer to manually derived measurements in other samples have demonstrated high degrees of agreement between the approaches,18 with correlations as high as 0.82 for hippocampal volume estimates.20 The observed hippocampal volumes did not differ across the scanning sites. An estimate of total intracranial volume (ICV) was also derived from the FreeSurfer atlas scaling factor on the basis of the transformation of the full brain mask into atlas space.21 Estimated ICV was used to control the hippocampal measures for differences in head size.19
APOE genotyping.
APOE genotype was determined from blood samples using established methods.22,23 All genotypes were independently determined twice by laboratory personnel at the VA Puget Sound Healthcare System who were blind to the genotype and the identity of the cotwin. Of the 375 participants utilized for the present analyses, 2 (0.5%) possessed a 2/2 genotype, 58 (15.5%) were 2/3, 16 (4.2%) were 2/4, 220 (58.7%) were 3/3, 70 (18.7%) were 3/4, and 9 (2.4%) were 4/4. These rates are roughly equivalent to those found in the general population.24 Participants with at least 1 copy of the ε4 allele were classified as being ε4 positive (ε4+); all other participants were classified as ε4 negative (ε4−).
Health and medical data.
Due to the well-established relationship between low testosterone, increased body mass index (BMI), and increased health problems,25–27 we included measures of BMI and overall health as additional covariates. Each participant underwent a medical history interview during which they were asked whether a physician had diagnosed them with any of 48 medical conditions (e.g., hypertension, high cholesterol, diabetes). The total number of conditions endorsed was utilized as a proxy for overall health.
Statistical analysis.
Data were analyzed using a multilevel, mixed linear model (SAS Proc Mixed, SAS version 9.2), which allowed for the utilization of all available data while adjusting for the nonindependence of the observations. Given the natural clustering of twin data, each member of a twin pair was identified by a unique ID number as well as a twin-pair specific number, referred to as the family ID. Similarly, each assay batch was assigned a unique ID number so as to control for the clustering imposed by the laboratory processing. Both family ID and batch ID were entered as random effects in the model. Although the present sample was collected as part of a twin study, the analyses performed were not traditional twin analyses. Zygosity was not utilized as a covariate, and hippocampal volumes and testosterone levels did not differ between monozygotic and dizygotic groups.
Analyses examined the effects of APOE genotype, testosterone, and their interaction on the left and right hippocampal volumes. The statistical model included estimated ICV, age, and handedness as initial covariates. Significant relationships were evaluated using the type III test of fixed effects, controlling for all other elements of the model. Due to the fact that clinical guidelines for the assessment of testosterone levels suggest sampling during the morning hours, generally between 8 and 10 am,28 we utilized the average 10 am sample from all 3 collection days as our primary hormone measure. We also examined the average testosterone level for all samples from the 3 collection days. Because the VETSA sample is a relatively young, nonpatient sample, we believed that testosterone effects were most likely to be observed toward one end of the distribution. Therefore, we utilized a statistical definition such that participants were classified as having low testosterone if their levels were 1 SD or more below the mean, while all other participants were classified as normal. The 10 am value is most comparable to the measures used in large epidemiologic studies. Our cutoff for this time point of 59.8 pg/mL is similar to or more conservative than cutpoints for hypogonadism based on free testosterone in large epidemiologic studies.29,30
RESULTS
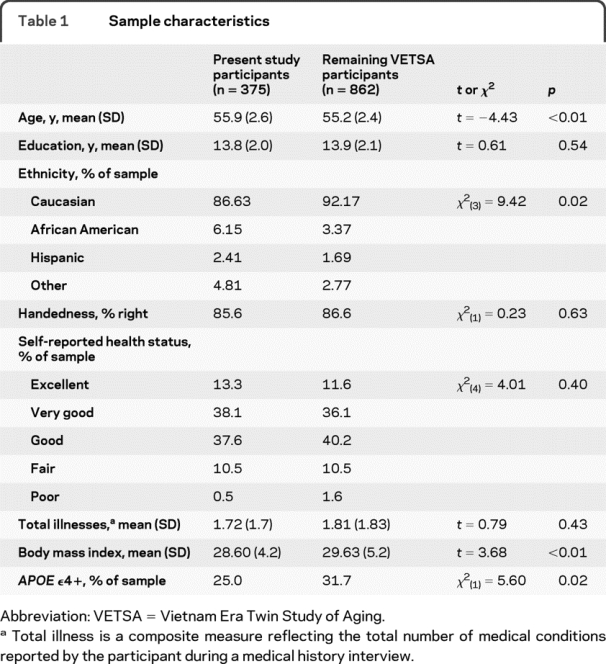
Demographic and other descriptive data are presented in table 1. The present sample was marginally older than the remaining VETSA participants (average age 55.9 vs 55.2), had a slightly smaller proportion of Caucasians, and had a lower average BMI. The present sample also had a lower prevalence of the APOE ε4 allele relative to the remaining VETSA participants: 25.0% vs 31.7%. The majority of the participants (89%) described their overall health as good or better.
Table 1 Sample characteristics
The average 10 am testosterone level across the sampling days was 93.7 pg/mL (SD = 33.9). The correlations between days ranged from 0.34 to 0.58 (Cronbach α = 0.69). The average testosterone level across all sampling days was 95.0 pg/mL (SD = 28.5). In contrast to the 10 am measures, the correlations between the daily averages were noticeably higher, ranging from 0.67 to 0.78 (Cronbach α = 0.88). Dichotomizing the 10 am testosterone sample resulted in 54 individuals in the low testosterone group and 321 in the normal testosterone group. For the daily average measurement, 60 participants were classified as having low testosterone. The tetrachoric correlation between the 2 dichotomous testosterone measures was 0.81 (95% CI 0.70–0.89). Participants who were classified as having low testosterone were not older than their normal testosterone counterparts, nor did they differ with respect to the prevalence of the ε4 allele. There were no significant differences in the proportion of ε4+ and ε4− participants in the low or normal testosterone groups.
10 am testosterone measures.
Results from the mixed linear models are presented in table 2. For the left hippocampus, a significant main effect of APOE genotype was observed, as was a significant interaction effect between APOE genotype and testosterone. There was no significant main effect of testosterone. As can be seen in the figure, participants with at least one copy of the ε4 allele and low testosterone had the smallest left hippocampal volumes of the 4 groups, although they were only significantly different from their ε4− low testosterone counterparts. The ε4− participants with low testosterone levels had significantly larger left hippocampal volumes relative to all other groups. Removal of the interaction from the model resulted in a loss of the significant main effect of APOE genotype, suggesting the presence of a suppression effect in which the interaction increases the predictive effect of the genotype.31 The same pattern of results was observed for the right hippocampus. After including BMI and total illnesses as additional covariates in the models, all previously significant main and interaction effects remained significant.
Table 2 Mixed linear model results
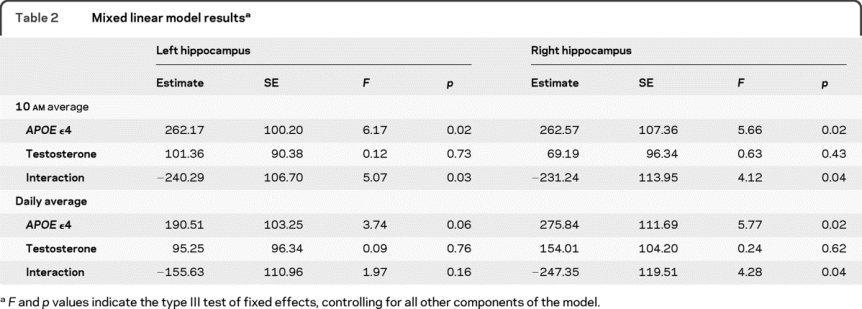
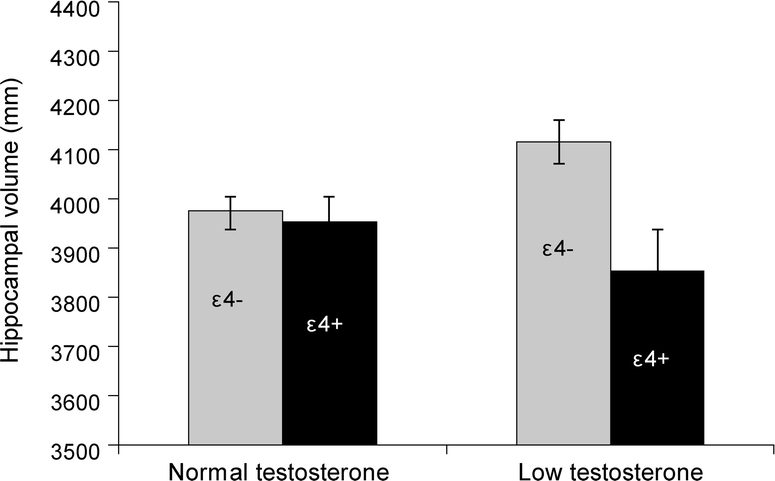
Figure Left hippocampal volume as a function of APOE and 10 am testosterone levels
The total number of participants in each group is as follows: normal testosterone/ε4 negative n = 241, normal testosterone/ε4 positive n = 80, low testosterone/ε4 negative n = 39, low testosterone/ε4 positive n = 15.
Daily average testosterone measures.
When the daily average testosterone measure was utilized, effects for the right hippocampus were consistent with the 10 am measure; there was a significant main effect of APOE genotype and a significant interaction with testosterone. However, the previously observed main and interaction effects for the left hippocampus were less pronounced. The effect of APOE genotype on left hippocampal volume was nearly significant (p = 0.06), while the interaction with testosterone was not significant. As with the previous analyses, ε4+ participants with low testosterone levels had smaller hippocampal volumes relative to the other groups; however, only the comparison with the ε4− low testosterone group was significant. The main effect of APOE genotype once again became nonsignificant when the interaction was removed from the model.
DISCUSSION
We observed a significant interaction between APOE genotype and testosterone with a patterning of group differences that was consistent with our initial hypothesis. Participants with the combination of at least one copy of the ε4 allele and low testosterone possessed the smallest hippocampal volumes, although comparisons relative to the normal testosterone groups were not statistically significant. Testosterone alone had no impact on hippocampal volume, yet the main effect of APOE genotype did become significant after the APOE-by-testosterone interaction was included in the model. Had our examination of the relationship between APOE genotype and hippocampal volume not included the interaction with testosterone, we would have concluded that APOE genotype does not influence hippocampal volume in middle-aged adults without dementia. While there have been previous findings of APOE-by-testosterone interactions, to our knowledge this is the first demonstration of such an effect on brain structure in middle-aged adults without dementia.
Interestingly, individuals who lacked the ε4 allele and had low testosterone were found to have significantly larger hippocampal volumes relative to all other groups. This result, while not predicted by our a priori hypothesis, is nevertheless consistent with a number of animal studies in which the relationship between APOE genotype, testosterone, ARs, and the hippocampus have been shown to be highly complex. For instance, expression of the testicular feminization mutation in mice, which truncates the N-terminal activation domain and renders the AR nonfunctional, has been found to attenuate the detrimental effect of APOE ε4 on some but not all hippocampal-dependent tasks.32 Comparable effects have been observed through the use of castration with no alteration of the AR.33 Treatment of castrated rats with a selective AR modulator has also been shown to restore AR expression within the brain and improve hippocampal-dependent learning and memory despite the absence of circulating testosterone levels.34 These results suggest that the interaction of circulating testosterone levels and the APOE genotype may be affected by other factors such as variations of the AR or modulators of the AR within the hippocampus. Thus, our findings may suggest the presence of 2 low testosterone subgroups.
It remains unclear what the exact biologic mechanism is that underlies the present results. One possibility is that low testosterone levels, occurring independently of the APOE genotype, result in fewer or less efficient ARs within the hippocampus, leaving the region more susceptible to the effects of the ε4 allele. Such a mechanism could be viewed as supportive of the lack of a main effect for testosterone we observed, as well as the presence of the interaction increasing the predictive effect of APOE ε4. Alternatively, it is possible that the APOE ε4 genotype may influence the hypothalamus, and as a result alter the function of the hypothalamic-pituitary axis. This could then result in lower testosterone levels via disrupted innervations of the testes, as well as differences in hippocampal volume as a result of changes in hormones such as cortisol. If this later scenario were the case, however, we would have likely observed a significantly higher prevalence of ε4+ participants in our low testosterone group relative to the normal testosterone group. No such differences were observed in the present sample.
The observed interaction between APOE and testosterone likely represents a form of gene-by-environment or a gene-by-gene interaction. APOE and testosterone are linked through a common metabolic pathway, the catabolism of the constituent cholesterol esters of lipoproteins, and the associated gene pathways offer a number of possible gene–gene interactions. It is important to note, however, that the level of free testosterone is not determined solely by genes, and that in adult men environmental influences account for roughly 50% of the variance in testosterone level.35,36 Further examination of this relationship will need to establish candidate genetic and environmental factors that influence testosterone levels in order to determine if they indeed interact with APOE genotype.
There are some potential limitations of this study. First, the all-male composition of this sample limits our ability to generalize these findings to women. To a large extent, research on estrogen loss in women parallels the work that has been done on testosterone in men, establishing connections with APOE,37 as well as demonstrating effects on hippocampal structure and gene expression.38,39 In addition to estrogen, women also experience late-life declines in testosterone.40 The distinct pattern of age-related changes of testosterone and estrogen in men and women may complicate any potential comparison; nevertheless, the possibility of similar interactions in women warrants future investigation. Second, even with our very large MRI sample, there were only a small number of participants with both an ε4 allele and low testosterone. Similarly, we were unable to examine ε4 dose effects or the effects of other APOE alleles (e.g., ε2). Thus, it will be important to see if our results are replicated in independent samples. Finally, the present analyses were cross-sectional; thus, we are unable to determine if the observed interactions are the result of age-related changes in hormone levels or represent longstanding relationships.
AUTHOR CONTRIBUTIONS
Statistical analysis was conducted by Dr. Panizzon.
ACKNOWLEDGMENT
Assistance was provided by the VA Cooperative Studies Program; Department of Defense; National Personnel Records Center, National Archives and Records Administration; the Internal Revenue Service; NIH; National Opinion Research Center; National Research Council, National Academy of Sciences; the Institute for Survey Research, Temple University; and Schulman, Ronca, & Bucuvalas, Inc. The authors thank the members of the Vietnam Era Twin Registry and their families for cooperation and participation.
DISCLOSURE
Dr. Panizzon has received fellowship support from the NIH/NIA (R01 AG018386, R01 AG018384, R01 AG022381, and R01 AG022982). Dr. Hauger reports no disclosures. Dr. Dale is a founder, holds equity in, and serves on the scientific advisory board of CorTechs Laboratories, Inc.; and receives research support from GE Healthcare, and the NIH (R01MH079752-04 [coinvestigator], P50MH081755-03 [Project PI], R01AG031224-02 [PI], R01EB009282-02 [coinvestigator], R01AG012674-12 [coinvestigator], P50AG005131-26 [coinvestigator], P50DA026306-01A1 [coinvestigator], RC2DA029475-01 [Co-PI], and R01AG022381-07 [coinvestigator]). Dr. Eaves receives research support from the NIH (NIAID 1UH2AI083263 [coinvestigator], NIDA U01 DA024413 [coinvestigator], NICHD R01 049685 [coinvestigator], R01 AG022381 [coinvestigator], NRSA T32 MH20030 [coinvestigator], NIMH R01 NIA 022982 [coinvestigator], NIDA R01 DA23668 [coinvestigator], NIA R01 AG18384 [coinvestigator]). Dr. Eyler receives research support from the NIH (MH083968-01 [PI], AG022381-07 [coinvestigator], MH091755-01 [coinvestigator], and MH036840-21 [coinvestigator]), UC San Diego Academic Senate, and from VA VISN 22 Mental Illness Research Education and Clinical Center. Dr. Fischl has served as a consultant for Cephalon, Inc.; and receives research support from the NIH (P41-RR14075 [Project Leader], U24 RR021382 [PI of Subcontract], R01EB006758 [PI], R01AG02238 [PI of subcontract], and R01NS052585 [PI]), and from the Ellison Medical Foundation. Dr. Fennema-Notestine receives research support from the NIH (R21 NS069355 [PI], N01 MH22005 [coinvestigator], P30 MH062512 [coinvestigator], P50 DA026306 [coinvestigator], R01 MH079752 [coinvestigator], R01 AG031224 [coinvestigator], R01 AG022381 [coinvestigator], U24 RR021992 [coinvestigator], R01 MH084796 [coinvestigator], U24 RR21382 [coinvestigator], U24 RR019701 [coinvestigator], and R01 AG024506 [coinvestigator]), the US Department of Veterans Affairs, and the Alzheimer's Association. Dr. Franz receives research support from the NIH (NIA R01 AG018386 [coinvestigator], R01 AG022381 [coinvestigator], and R01 AG022982 [coinvestigator]). Dr. Grant reports no disclosures. Dr. Jak receives research support from the NIH (NIA 5 RO1 AG12674 [coinvestigator] and NIMH 1R43AG032629-01A1 SBIR [coinvestigator]), the US Department of Veterans Affairs, and the Alzheimer's Association. Dr. Jacobson serves as a consultant for the NIH (PhenX project); served as Editor for Special Issue of Behavior Genetics; and receives research from the NIH (1DP2 OD003021 [PI], NIMH 5R01 MH058354 [PI of subcontract], NIA 1R01 AG018386 [PI of subcontract], NIMH 1R01 MH080109 [coinvestigator], and NIMH/NIA 5R01 AG022982 [PI of subcontract]). Dr. Lyons serves as an Associate Editor of Behavior Genetics; and receives research support from the NIH (NIA 5R01AG018384 [PI]). Dr. Mendoza receives research support from the NIH (NCRR RR00169 [Staff Scientist], NCRR RR019970 [coinvestigator], and NICHD R01 HD053555 [Co-PI]) and from the California National Primate Research Center (NIH). Dr. Neale receives research support from the NIH (DA-26119 [PI], DA-18673 [PI], and AG-22381 [PI subcontract]). Dr. Prom-Wormley has received support from the NIH/NIMH (Training Grant MH-20030). Dr. Seidman serves as an Associate Editor of Neuropsychology; and receives research support from the NIH (NIMH 1 P50 MH080272-01 [PI clinical core], NIMH 1 U01 MH081928-01A1 [PI], NIMH 5R01 MH56956 [coinvestigator], NIMH 4RO1 MH040799 [coinvestigator], NIMH R21 MH083205 [coinvestigator], NIMH 1 RO1 MH078113-01A2 [coinvestigator], and 1R21 MH082235 [coinvestigator]), the Massachusetts Department of Mental Health, UCLA, and from the Sidney Baer Foundation. Dr. Tsuang serves as Editor-in-Chief of the American Journal Medical Genetics Part B, Neuropsychiatric Genetics; is an inventor on patents re: Biomarkers for Psychosis; receives royalties from the publication of Textbook of Psychiatric Epidemiology, 2nd ed. (Wiley-Liss, 2002); and receives research support from the NIH (R21 MH85240 [PI], R01 MH085560 [PI], and R01 MH081861 [PI]). Dr. Xian receives research support from the NIH (NIA R01 AG18386-5 [coinvestigator], NIDA R01 DA020810 [PI], NHGRI 5 U01 HG04422-2 [coinvestigator], and NIDA 2 R01 DA014262-06 [coinvestigator]). Dr. Kremen serves on the editorial board of Neuropsychology Review; is an inventor on a patent re: Detection and Validation of Biomarkers for Neuropsychiatric Disorders (NeuroMark Genomics, Inc.); and serves as a grant reviewer on study section for the US Veterans Administration.
Address correspondence and reprint requests to Dr. Matthew S. Panizzon, Department of Psychiatry, University of California, San Diego, 9500 Gilman Drive (MC 0738), La Jolla, CA 9293-0738 mspanizz@ucsd.edu
Study funding: Supported by the NIH/NIA (R01 AG018386, RO1 AG018384, RO1 AG022381, and RO1 AG022982) (VETSA project).
Presented in part at the annual meeting of the International Society of Psychoneuroendocrinology, San Francisco, CA, July 2009. The US Department of Veterans Affairs has provided support for the development and maintenance of the Vietnam Era Twin Registry.
Disclosure: Author disclosures are provided at the end of the article.
Received January 28, 2010. Accepted in final form May 12, 2010.
REFERENCES
- 1. Saunders AM Strittmatter WJ Schmechel D et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 2. Cherbuin N Leach LS Christensen H Anstey KJ Neuroimaging and APOE genotype: a systematic qualitative review. Dement Geriatr Cogn Disord. 2007;24:348–362. doi: 10.1159/000109150. [DOI] [PubMed] [Google Scholar]
- 3.Wisdom NM, Callahan JL, Hawkins KA. The effects of apolipoprotein E on non-impaired cognitive functioning: a meta-analysis. Neurobiol Aging (in press 2010). [DOI] [PubMed]
- 4. Moore JH Williams SM New strategies for identifying gene-gene interactions in hypertension. Ann Med. 2002;34:88–95. doi: 10.1080/07853890252953473. [DOI] [PubMed] [Google Scholar]
- 5. Moffat SD Zonderman AB Metter EJ et al. Free testosterone and risk for Alzheimer disease in older men. Neurology. 2004;62:188–193. doi: 10.1212/wnl.62.2.188. [DOI] [PubMed] [Google Scholar]
- 6. Chu LW Tam S Lee PW et al. Bioavailable testosterone is associated with a reduced risk of amnestic mild cognitive impairment in older men. Clin Endocrinol. 2008;68:589–598. doi: 10.1111/j.1365-2265.2007.03094.x. [DOI] [PubMed] [Google Scholar]
- 7. Rosario ER Pike CJ Androgen regulation of beta-amyloid protein and the risk of Alzheimer's disease. Brain Res Rev. 2008;57:444–453. doi: 10.1016/j.brainresrev.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moffat SD Resnick SM Long-term measures of free testosterone predict regional cerebral blood flow patterns in elderly men. Neurobiol Aging. 2007;28:914–920. doi: 10.1016/j.neurobiolaging.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 9. Raber J Bongers G LeFevour A Buttini M Mucke L Androgens protect against apolipoprotein E4-induced cognitive deficits. J Neurosci. 2002;22:5204–5209. doi: 10.1523/JNEUROSCI.22-12-05204.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raber J Androgens, apoE, and Alzheimer's disease. Science Aging Knowl Environ. 2004;2004:1–11. doi: 10.1126/sageke.2004.11.re2. [DOI] [PubMed] [Google Scholar]
- 11. Hogervorst E Lehmann DJ Warden DR McBroom J Smith AD Apolipoprotein E epsilon4 and testosterone interact in the risk of Alzheimer's disease in men. Int J Geriatr Psychiatry. 2002;17:938–940. doi: 10.1002/gps.714. [DOI] [PubMed] [Google Scholar]
- 12. Hyman BT Damasio H Damasio AR Van Hoesen GW Alzheimer's disease. Annu Rev Public Health. 1989;10:115–140. doi: 10.1146/annurev.pu.10.050189.000555. [DOI] [PubMed] [Google Scholar]
- 13. Beyenburg S Watzka M Clusmann H et al. Androgen receptor mRNA expression in the human hippocampus. Neurosci Lett. 2000;294:25–28. doi: 10.1016/s0304-3940(00)01542-1. [DOI] [PubMed] [Google Scholar]
- 14. Kremen WS Thompson-Brenner H Leung YJ et al. Genes, environment, and time: The Vietnam Era Twin Study of Aging (VETSA). Twin Res Hum Genet. 2006;9:1009–1022. doi: 10.1375/183242706779462750. [DOI] [PubMed] [Google Scholar]
- 15. Goldberg J Curran B Vitek ME Henderson WG Boyko EJ The Vietnam Era Twin Registry. Twin Res Hum Genet. 2002;5:476–481. doi: 10.1375/136905202320906318. [DOI] [PubMed] [Google Scholar]
- 16.Centers for Disease Control and Prevention. Health data for all ages. Available at: http://www.cdc.gov/nchs/health_data_for_all_ages.htm. Accessed April 20, 2007.
- 17. Granger DA Schwartz EB Booth A Arentz M Salivary testosterone determination in studies of child health and development. Horm Behav. 1999;35:18–27. doi: 10.1006/hbeh.1998.1492. [DOI] [PubMed] [Google Scholar]
- 18. Fischl B Salat DH Busa E et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 19. Kremen WS Prom-Wormley E Panizzon MS et al. Genetic and environmental influences on the size of specific brain regions in midlife: the VETSA MRI study. Neuroimage. 2010;49:1213–1223. doi: 10.1016/j.neuroimage.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morey RA Petty CM Xu Y et al. A comparison of automated segmentation and manual tracing for quantifying hippocampal and amygdala volumes. Neuroimage. 2009;45:855–866. doi: 10.1016/j.neuroimage.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buckner RL Head D Parker J et al. A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas-based head size normalization: reliability and validation against manual measurement of total intracranial volume. Neuroimage. 2004;23:724–738. doi: 10.1016/j.neuroimage.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 22. Emi M Wu LL Robertson MA et al. Genotyping and sequence analysis of apolipoprotein E isoforms. Genomics. 1988;3:373–379. doi: 10.1016/0888-7543(88)90130-9. [DOI] [PubMed] [Google Scholar]
- 23. Hixson JE Vernier DT Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res. 1990;31:545–548. [PubMed] [Google Scholar]
- 24. Zannis VI Kardassis D Zanni EE Genetic mutations affecting human lipoproteins, their receptors, and their enzymes. Adv Hum Genet. 1993;21:145–319. doi: 10.1007/978-1-4615-3010-7_3. [DOI] [PubMed] [Google Scholar]
- 25. Barrett-Connor EL Testosterone and risk factors for cardiovascular disease in men. Diabetes Metab. 1995;21:156–161. [PubMed] [Google Scholar]
- 26. Rosmond R Wallerius S Wanger P Martin L Holm G Björntorp P A 5-year follow-up study of disease incidence in men with an abnormal hormone pattern. J Intern Med. 2003;254:386–390. doi: 10.1046/j.1365-2796.2003.01205.x. [DOI] [PubMed] [Google Scholar]
- 27. Travison TG Araujo AB Kupelian V O'Donnell AB McKinlay JB The relative contributions of aging, health, and lifestyle factors to serum testosterone decline in men. J Clin Endocrinol Metab. 2007;92:549–555. doi: 10.1210/jc.2006-1859. [DOI] [PubMed] [Google Scholar]
- 28. Petak SM Nankin HR Spark RF Swerdloff RS Rodriguez-Rigau LJ American Association of Clinical Endocrinologists Medical Guidelines for clinical practice for the evaluation and treatment of hypogonadism in adult male patients: 2002 update. Endocrine Pract. 2002;8:440–456. [PubMed] [Google Scholar]
- 29. Araujo AB O'Donnell AB Brambilla DJ et al. Prevalence and incidence of androgen deficiency in middle-aged and older men: estimates from the Massachusetts Male Aging Study. J Clin Endocrinol Metab. 2004;89:5920–5926. doi: 10.1210/jc.2003-031719. [DOI] [PubMed] [Google Scholar]
- 30. Mulligan T Frick MF Zuraw QC Stemhagen A McWhirter C Prevalence of hypogonadism in males aged at least 45 years: the HIM study. Int J Clin Pract. 2006;60:762–769. doi: 10.1111/j.1742-1241.2006.00992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen J, Cohen P. Applied Multiple Regression/Correlation Analysis for the Behavioral Sciences, 2nd ed. Hillsdale, NJ: Erlbaum Associates; 1983. [Google Scholar]
- 32. Rizk-Jackson A Robertson J Raber J Tfm-AR modulates the effects of ApoE4 on cognition. J Neurochem. 2008;105:63–67. doi: 10.1111/j.1471-4159.2007.05092.x. [DOI] [PubMed] [Google Scholar]
- 33. Pfankuch T Rizk A Olsen R Poage C Raber J Role of circulating androgen levels in effects of apoE4 on cognitive function. Brain Res. 2005;1053:88–96. doi: 10.1016/j.brainres.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 34. Acevedo S Gardell L Bradley SR Piu F Raber J Selective androgen receptor modulators antagonize apolipoprotein E4 induced cognitive impairments. Lett Drug Design Discov. 2008;5:271–276. [Google Scholar]
- 35. Ring HZ Lessov CN Reed T et al. Heritability of plasma sex hormones and hormone binding globulin in adult male twins. J Endocrinol Metab. 2005;90:3653–3658. doi: 10.1210/jc.2004-1025. [DOI] [PubMed] [Google Scholar]
- 36. Kuijper EA Lambalk CB Boomsma DI et al. Heritability of reproductive hormones in adult male twins. Hum Reprod. 2007;22:2153–2159. doi: 10.1093/humrep/dem145. [DOI] [PubMed] [Google Scholar]
- 37. MacLusky NJ Estrogen and Alzheimer's disease: the apolipoprotein connection. Endocrinology. 2004;145:3062–3064. doi: 10.1210/en.2004-0427. [DOI] [PubMed] [Google Scholar]
- 38. Aenlle KK Kumar A Cui L Jackson TC Foster TC Estrogen effects on cognition and hippocampal transcription in middle-aged mice. Neurobiol Aging. 2009;30:932–945. doi: 10.1016/j.neurobiolaging.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen JR Yan YT Wang TJ Chen LJ Wang YJ Tseng GF Gonadal hormones modulate the dendritic spine densities of primary cortical pyramidal neurons in adult female rat. Cereb Cortex. 2009;19:2719–2927. doi: 10.1093/cercor/bhp048. [DOI] [PubMed] [Google Scholar]
- 40. Bachmann G Bancroft J Braunstein G et al. Female androgen insufficiency: the Princeton consensus statement on definition, classification, and assessment. Fertil Steril. 2002;77:660–665. doi: 10.1016/s0015-0282(02)02969-2. [DOI] [PubMed] [Google Scholar]


