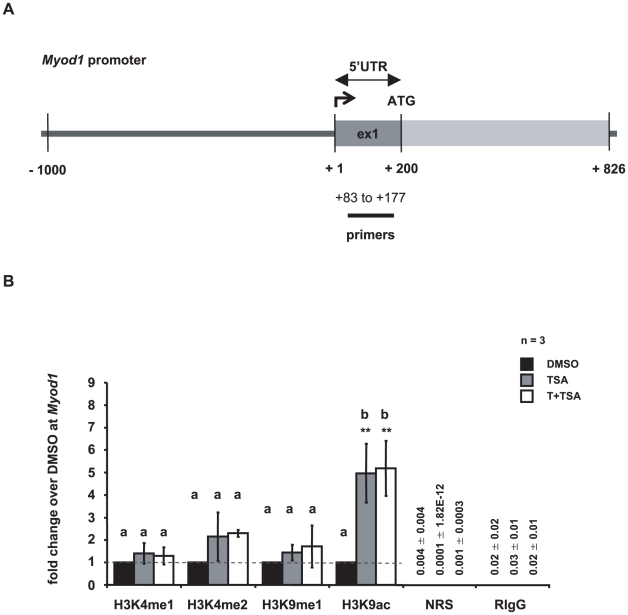
Figure 5. Histone H3 methylation at Myod1 promoter is not affected by tranylcypromine and trichostatin A treatment.
(A) Schematic representation of the Myod1 promoter. Numbers depict the positions of primers used for ChIP-qPCR relative to the corresponding transcriptional start site. (B) ChIP analyses followed by qPCR reveal changes in the epigenetic landscape of the Myod1 promoter upon treatment. Despite the increase in H3K9 acetylation, Myod1 is not expressed in untreated GC-1 cells nor induced as a consequence of treatment, as shown in Table S4. Depicted are histone H3 methylation and acetylation levels in GC-1 cells that have been cultured in the presence of dimethylsulphoxide (DMSO, control, black bars), or trichostatin A (TSA, grey bars), or both inhibitors, tranylcypromine and TSA (T+TSA, white bars). Signals of target DNA received from DMSO control cells served as base and were defined as one. Fold change indicates 2∧-(ΔΔCT) of target DNA in treated cells over 2∧-(ΔΔCT) of target DNA in DMSO controls (Y-axis). Epigenetic modifications are depicted on the X-axis. Data are expressed as mean ± Stdev; *p<0.05, **p<0.01. Means with different letters are significantly different. N indicates the number of independent experiments, and per experiment each sample and the corresponding negative controls were run in triplicates.