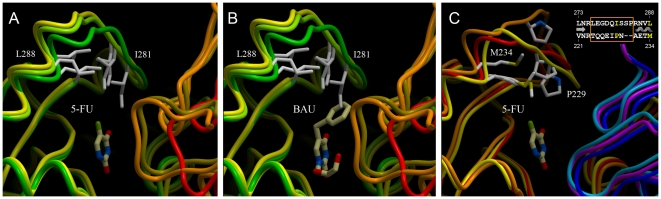
Figure 3. Conformational dynamics of a hUPP1 active site loop.
(A) Comparison of the structure of the loop lining the back of the hUPP1 active site when bound to 5-FU (green), BAU (lime), or ligand-free (yellow), reveals that this region is somewhat mobile and able to close around substrate upon its binding. (B) Overlay of the BAU molecule with the known structures of hUPP1 shows that the benzyl moiety of this inhibitor displaces Ile281 from its normal substrate binding position to accommodate the extra bulkiness of this molecule. It is notable how similar the BAU-bound and ligand-free conformations of hUPP1 are, suggesting that BAU fits the naturally occurring structure of the protein in the absence of substrate. (C) While the new structure of hUPP1 reveals some degree of flexibility in the back-side active site loop, the conformational range of this region of hUPP1 is substantially less than that of the equivalent part of E. coli UPP, which closes more tightly when bound to 5-FU (yellow) and opens wider in the absence of ligand (orange) when compared with its BAU-inhibited structure (red). The increased rigidity of the human enzyme is likely due to the insertion of two additional residues into this loop region, including a proline (inset).