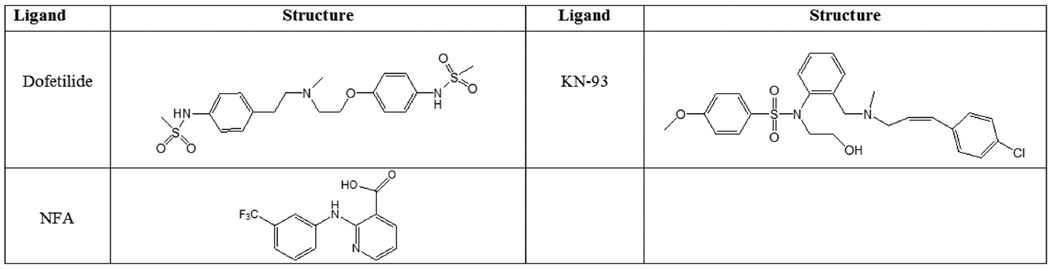
Figure 2. Chemical Structures of the ligands used for molecular docking studies.
Evaluation of constructed hERG1 model and elucidation of possible binding sites have been performed by molecular docking simulations. Dofetilide and KN-93 were used as representatives of hERG1 blockers and NFA was used as an activator.