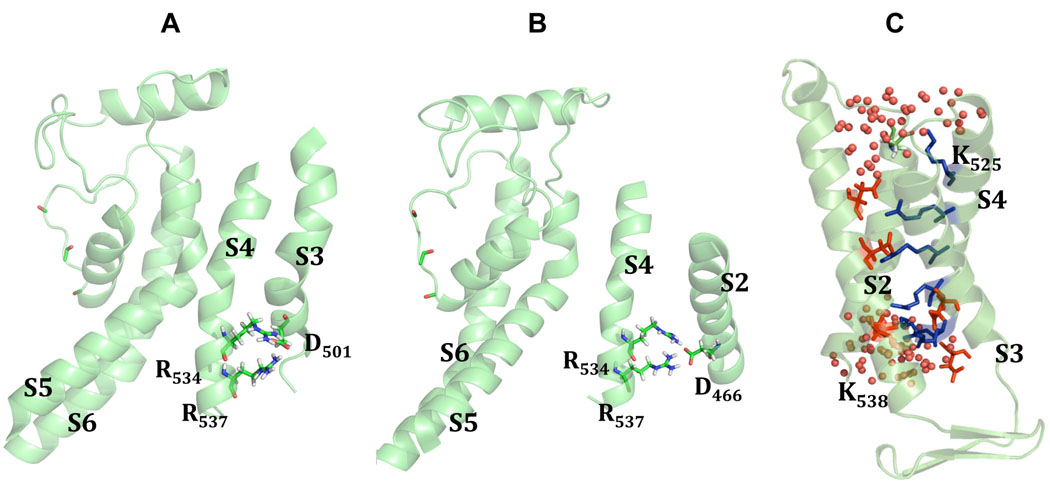
Figure 5. Critical Interactions in VS domain in herg+3 model.
The analysis of interaction energies for R534/R537 suggests that these two residues may form bifurcating salt-bridges with D501 (A) and D466 (B). Panel C shows water accessibility of the S4 helix averaged from MD simulations of the herg+3 model. Two lysines (K525 and K538) were found to be exposed to the lipid-water interface, while other four arginines (R528, R531, R534, R537) are buried inside of voltage-sensor bundle forming stable contacts to negatively charged residues from S1 and S2 TM helices.