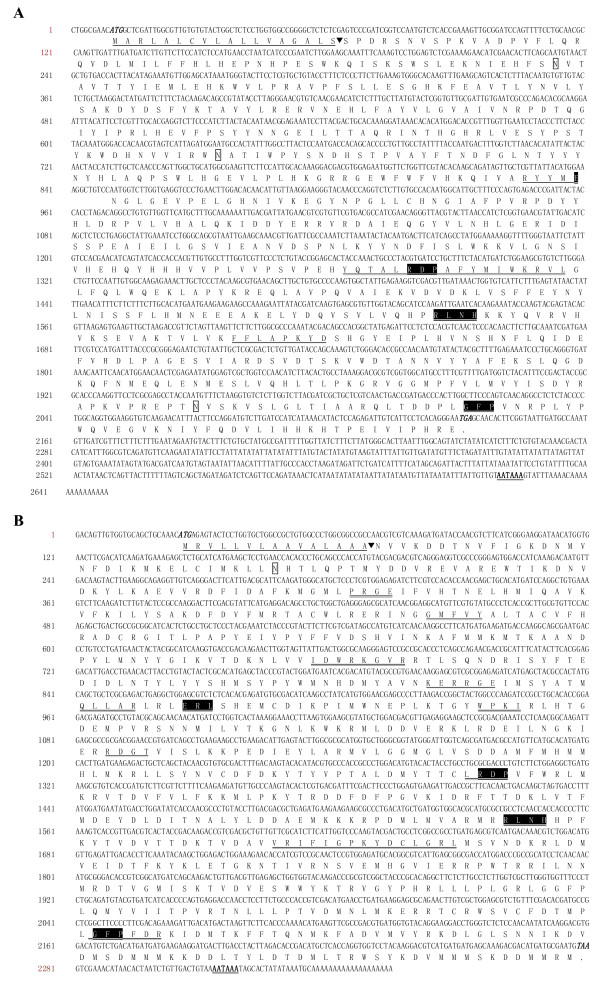
Figure 1.
Nucleotide and deduced amino acid sequences for S. exigua Hex and SP1 cDNAs. Italic and bold nucleotides indicate the start and stop codons, respectively. The termination signal AATAAA is bold and underlined. The hemocyanin group of motifs (or signature motifs) (amino acid residues ERL, RDP, RLNH and GFP) are shaded in black. (A) SeHex cDNA sequence analysis. Underlined amino acid residues (1-18) and the arrowhead represent the signal peptide and putative cleavage site, respectively. Regions that are highly conserved in lepidopteran Hex genes are double underlined. Potential N-glycosylation sites (residues 75, 209, 479 and 647) are boxed. The nucleotide sequence reported in this paper has been submitted to GenBank (accession number EF646282). (B) SeSP1 cDNA sequence analysis. Underlined amino acid residues (1-15) and the arrowhead represent the signal peptide and putative cleavage site, respectively. Regions that are highly conserved in lepidopteran SP1 genes are double underlined. The potential N-glycosylation site (residue 47) is boxed. The nucleotide sequence reported in this paper has been submitted to GenBank (accession number EU259816).