Abstract
The cofactors of the Mo- and V-nitrogenases (i.e., FeMoco and FeVco) are homologous metal centers with distinct catalytic properties. So far, there has been only one report on the isolation of FeVco from Azotobacter chroocucum. However, this isolated FeVco species did not carry the full substrate-reducing capacity, as it is unable to restore the N2-reducing ability of the cofactor-deficient MoFe protein. Here, we report the isolation and characterization of a fully active species of FeVco from Azotobacter vinelandii. Our metal and activity analyses show that FeVco has been extracted intact, carrying with it the characteristic capacity to reduce C2H2 to C2H6 and, perhaps even more importantly, the ability to reduce N2 to NH3. Moreover, our EPR and XAS/EXAFS investigations indicate that FeVco is similar to, yet distinct from FeMoco in electronic properties and structural topology, which could account for the differences in the reactivity of the two cofactors. The outcome of this study not only permits the proposal of the first EXAFS-based structural model of the isolated FeVco, but also lays a foundation for future catalytic and structural investigations of this unique metallocluster.
Keywords: Nitrogenase, FeVco, FeMoco
1. Introduction
Nitrogenase catalyzes the nucleotide-dependent reduction of dinitrogen (N2) to ammonia (NH3).1 The “conventional” molybdenum (Mo)-nitrogenase is a binary system comprising (i) the iron (Fe) protein (encoded by nifH), an α2-dimer that contains a [Fe4S4] cluster at the subunit interface and a MgATP binding site within each subunit; and (ii) the nifDK-encoded molybdenum-iron (MoFe) protein (encoded by nifDK), an α2β2-tetramer that contains a P-cluster ([Fe8S7]) at each α/β subunit interface and a FeMoco ([MoFe7S9X-homocitrate], where X = C, N or O) within each α subunit (Figure 1).2 Likewise, the “alternative” vanadium (V)-nitrogenase is a two-component system comprising (i) the Fe protein (encoded by vnfH), an α2-dimer that contains a [Fe4S4] cluster; and (ii) the vanadium-iron (VFe) protein (encoded by vnfDGK), an α2β2δ4-octamer that contains a P-cluster and a FeVco in each half of the protein.3,4 The two nitrogenases share a great deal of homology with regards to both the primary sequences of their component proteins and the types of metal centers they contain. In particular, their active cofactors (i.e., FeMoco and FeVco), which provide the sites for substrate reduction, are believed to be structurally homologous.3 However, despite the considerable similarities in the structure and function of their cofactor centers, V-nitrogenase exhibits some unique catalytic features that are distinct from that of its Mo-counterpart,4 making it an attractive target for the investigation of the impact of heterometal on the physiochemical properties of nitrogenase cofactors. The development of an effective strategy for FeVco extraction, therefore, is highly desirable, as it affords the unique opportunity to study the FeVco without the interference of the P-cluster.
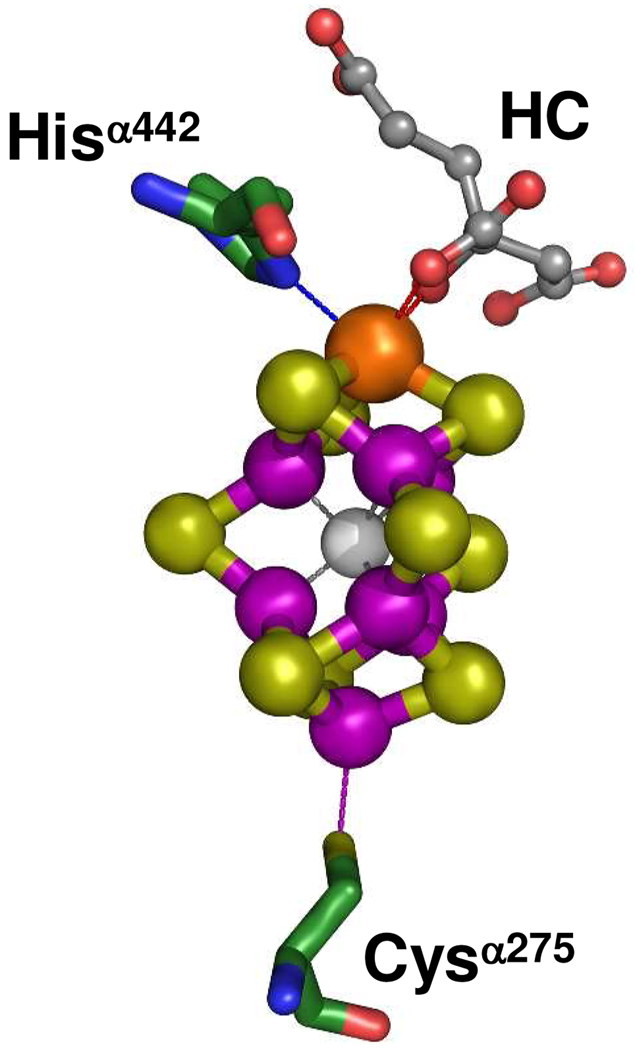
Figure 1.
Structure of the FeMoco of nitrogenase MoFe protein.2 The FeMoco is ligated to the MoFe protein by only two ligands: Cysα275 and Hisα442. In addition, its Mo atom is further coordinated by homocitrate (HC). The atoms of the metal cluster and protein ligands are colored as follows: Mo, orange; Fe, purple; S, lime; X (C, N or O), light gray; O, red; C, green; N, blue. The atoms of homocitrate (HC) are colored as follows: O, red; C, dark gray.
To date, there has been only one report on the extraction of FeVco from Azotobacter chroococum.5 While this initial study provided valuable insights into the enzymatic and EPR properties of the isolated FeVco, it left room for improvement, particularly with regard to (i) the weak EPR signal exhibited by the isolated FeVco; (ii) the low substrate-reducing activity carried by the isolated FeVco; and (iii) the inability of FeVco to activate the N2-reducing ability of the cofactor-deficient MoFe protein.5 Clearly, isolation of a catalytically fully-competent form of FeVco is the prerequisite for an accurate assessment of its physiochemical properties. One key factor that could impact the effectiveness of cofactor extraction is the quantity and quality of the starting material (i.e., the VFe protein). Recently, we have significantly improved the purification procedure of VFe protein, which enabled us to obtain a large quantity of this protein with a complete subunit composition.4 Here, we report the isolation and characterization of a fully active species of FeVco from Azotobacter vinelandii. Our metal and activity analyses show that FeVco has been extracted intact, carrying with it the characteristic capacity to reduce C2H2 to C2H6 and, perhaps even more importantly, the ability to reduce N2 to NH3. Moreover, our EPR and XAS/EXAFS investigation indicate that FeVco is similar to, yet distinct from FeMoco in electronic properties and structural topology, which could account for the differences in the reactivity of the two cofactors. The procedures established in this study not only facilitate the proposal of the first EXAFS-derived structural model of the isolated FeVco, but also provide a useful platform for further investigations of the physiochemical properties of this unique cluster.
2. Results and Discussion
Cluster Composition Analysis
The NMF-extracted FeVco is dark brown in color and contains V, Fe and acid-labile S6–9 in an approximate ratio of 1:7.4:7.2 (Table 1). In comparison, the NMF-extracted FeMoco is dark greenish-brown in color and contains Mo, Fe and acid-labile S in an approximate ratio of 1:7.3:7.1 (Table 1). The comparable Fe:V:S and Fe:Mo:S ratios suggest a similar cluster composition for FeVco and FeMoco. However, the slight variation in the colors of the two cofactor species points to a difference in their electronic properties.
Table 1.
Cluster composition (A) and reconstitution activities (B) of isolated cofactors. Substrate-reducing activities of cofactor-reconstituted ΔnifB MoFe protein were measured as described in Experimental Procedures. Percentages of substrate-reducing activities of FeVco-reconstituted ΔnifB MoFe protein relative to those of FeMoco-reconstituted ΔnifB MoFe protein are given in the parentheses.
| Cluster Type |
(A) Cluster Composition |
|||
|---|---|---|---|---|
| Fe/V ratio | S2−/V ratioa | Fe/Mo ratio | S2−/Mo ratioa | |
| FeVco | 7.4 ± 1.6 | 7.2 ± 1.8 | - | - |
| FeMoco | - | - | 7.3 ± 0.7 | 7.1 ± 0.5 |
| Cluster Type |
(B) Reconstitution Activitiesb |
||||
|---|---|---|---|---|---|
| C2H4-formation under C2H2/Ar |
C2H6-formation under C2H2/Ar |
H2-formation under Ar |
NH3-formation under N2 |
H2-formation from N2 |
|
| FeVco | 366 ± 27 (26) | 6 ± 1 | 363 ± 39 (25) | 166 ± 17 (23) | 174 ± 10 (80) |
| FeMoco | 1393 ± 101 (100) | < 0.3 | 1481 ± 110 (100) | 715 ± 82 (100) | 218 ± 13 (100) |
EPR and Activity Analysis
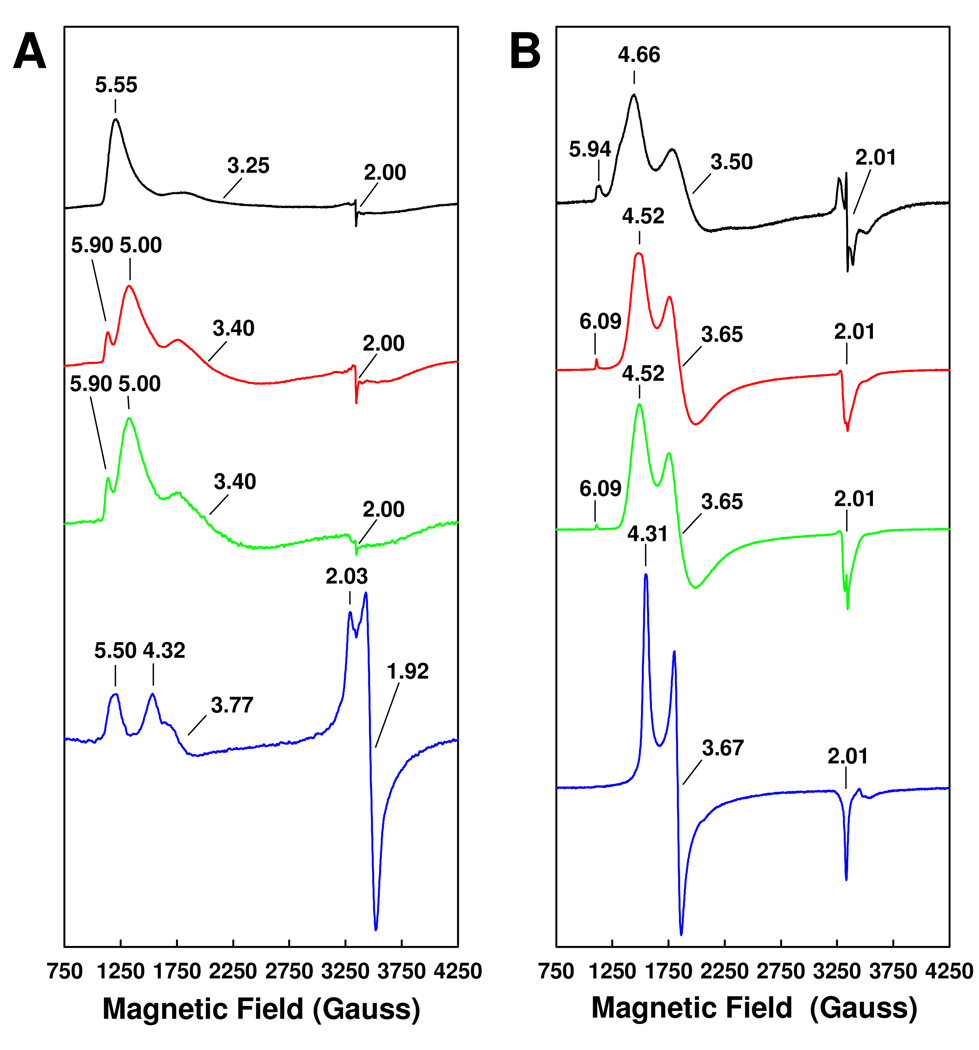
Upon extraction into NMF, both FeVco and FeMoco display three major features in their EPR spectra: FeVco shows a major feature at g = 5.55, a small (yet broad) feature at g = 3.25 and a minor feature at g = 2.00 (Figure 2A, black trace); whereas FeMoco exhibits a set of analogous features at g = 4.66, 3.50 and 2.01, respectively (Figure 2B, black trace). Both NMF-extracted cofactors (Figure 2, black traces) exhibit signals that are similar to, yet much broader than, those exhibited by their respective protein-bound counterparts (Figure 2, blue traces). Nevertheless, the signals of both isolated and protein-bound cofactors adopt a form that is characteristic of an S = 3/2 center,5,10 suggesting that the EPR active, S = 3/2 cofactor centers of both proteins have been successfully extracted into NMF. It is interesting to note that, once isolated, FeVco and FeMoco give rise to signals of a somewhat similar line shape (Figure 2, black traces), which is consistent with the presumed homology between the two cofactors.3 On the other hand, the isolated FeVco does possess spectroscopic properties that are distinct from those of the isolated FeMoco. Apart from having different g values, the signal of isolated FeVco is broader and less resolved than the signal of isolated FeMoco (Figure 2, black traces). Additionally, the isolated FeVco does not exhibit the extra g = 5.94 feature of the isolated FeMoco (Figure 2, black traces), which has been attributed to components of the Ms = 3/2 excited state of the S = 3/2 (Ms = 1/2 ground state) system.11 Finally, the g = 2.00 feature of the isolated FeVco is less dominant than the g = 2.01 feature of the isolated FeMoco (Figure 2, black traces). These spectral differences likely reflect the different electronic properties of the cofactors, as well as the differential interactions between the two cofactors and NMF.
Figure 2.
EPR properties of FeVco (A) and FeMoco (B). Shown are the EPR spectra of the cofactors in NMF (black traces), in NMF plus 10 mM thiophenol (red traces), in NMF plus 10 mM 1,4-benzene dithiol (green traces) or within the wild-type proteins (blue traces). The FeVco samples were measured at 6 K, whereas the FeMoco samples were measured at 15 K. The power and temperature at which the spectra were taken were determined on the basis of power- and temperature-dependent experiments, where the spectra were saturated in intensity and optimized for line-width. The (apparent) g values are indicated. All cofactor and protein samples contained equimolar V or Mo.
Following the addition of thiophenol and 1,4-benzene dithiol, the EPR signals of both isolated FeVco and isolated FeMoco (Figure 2, red and green traces) become sharper and assume line shapes similar to those of their respective protein-bound counterparts (Figure 2, blue traces). The g = 5.00 and g = 3.40 features of the isolated FeVco likely correspond to the g = 4.32 and g = 3.77 features of the VFe protein-associated FeVco; whereas the g = 4.52 and g = 3.65 features of the isolated FeMoco likely correspond to the g = 4.31 and g = 3.67 features of the MoFe protein-associated FeMoco. The additional features at g = 5.90 of the FeVco spectra (Figure 2A, red and green traces) and g = 6.09 of the FeMoco spectra (Figure 2B, red and green traces) may originate from the interactions of FeMoco and FeVco with the thiol groups in thiophenol and 1,4-benzene dithiol. More importantly, the ability of thiol groups to mimic the protein ligands for the isolated cofactors could account for the increased resemblance of thiol-treated cofactors to their protein-bound counterparts. Indeed, the effect of thiophenol addition to the NMF-extracted FeMoco has been studied by Mo K XANES and EXAFS analyses, which suggest a structural change induced by thiophenol binding that renders the isolated FeMoco in a conformation that resembles the protein-bound FeMoco more closely.10 While the cofactor-thiolate complexes are by no means identical to the protein-bound cofactors, the improved similarity of the isolated cofactors to their native counterparts in the presence of thiolate provides additional evidence that the cofactors have been isolated intact into NMF.
The integrity of the NMF-extracted cofactors is further demonstrated by the ability of isolated cofactors to reconstitute/activate the cofactor-deficient ΔnifB MoFe protein (Table 1). Consistent with the observation that VFe protein is less efficient than MoFe protein in catalysis, the ΔnifB MoFe protein is activated to a lesser degree by isolated FeVco than it is by isolated FeMoco (Table 1). The FeVco-reconstituted, “hybrid” MoFe protein is capable of generating both C2H4 and C2H6 as products of C2H2 reduction; in contrast, the FeMoco-reconstituted MoFe protein is only capable of reducing C2H2 to C2H4 (Table 1). Interestingly, the same discrepancy was observed between the C2H2-reducing profiles of the wild-type VFe and MoFe proteins.4 Under N2, the FeVco-reconstituted MoFe protein generates NH3 and H2 at a NH3/H2 ratio of 0.95, whereas the FeMoco-reconstituted MoFe protein forms these two products at a NH3/H2 ratio of 3.28 (Table 1). Again, the FeVco-reconstituted MoFe protein seems to mimic the wild-type VFe protein in the N2-reducing profile, as a similar shift of electron flow toward H2 evolution was also observed in the VFe protein-catalyzed reaction.4 Taken together, these observations point to a dominating effect of FeVco on the catalytic properties of the hybrid MoFe protein. On the other hand, while the wild-type VFe protein is very inefficient at C2H2 reduction,4 the FeVco-reconstituted MoFe protein reduces C2H2 as efficiently as H+ (under Ar) and N2 (Table 1), suggesting the impact of the immediate MoFe protein environment on the reactivity of the FeVco center.
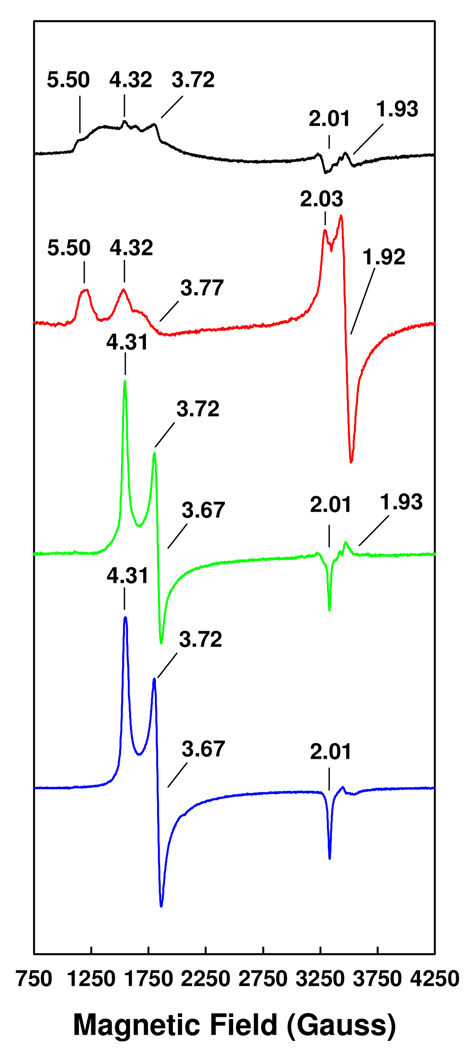
The mixed activity profile of the hybrid protein can be correlated with the “chimeric” EPR signal exhibited by its cofactor center. Like the VFe protein-bound FeVco (Figure 3, red trace), the MoFe protein-bound FeVco exhibits an S = 3/2 signal of a broad line shape, over which the characteristic features of FeVco at g = 5.50 and g = 4.32 can be observed (Figure 3, black trace). However, this unique S = 3/2 signal also displays the g = 3.72 feature (Figure 3, black trace) that is specific to the MoFe protein-associated FeMoco (Figure 3, blue trace). In contrast, the FeMoco signal can be completely restored upon the incorporation of isolated FeMoco into the ΔnifB MoFe protein (Figure 3, green trace), which is consistent with the observation that the homologously-reconstituted ΔnifB MoFe protein exhibits a substrate-reducing profile that is typical of the wild-type MoFe protein.
Figure 3.
EPR properties of the FeVco-reconstituted ΔnifB MoFe protein (black trace), the wild-type VFe protein (red trace), the FeMoco-reconstituted ΔnifB MoFe protein (green trace), and the wild-type MoFe protein (blue trace). Samples containing the FeVco were measured at 6 K, whereas those containing the FeMoco were measured at 15 K. The power and temperature at which the spectra were taken were determined on the basis of power- and temperature-dependent experiments, where the spectra were saturated in intensity and optimized for line-width. The (apparent) g values are indicated. All samples contained equimolar V or Mo.
XAS/EXAFS Analysis
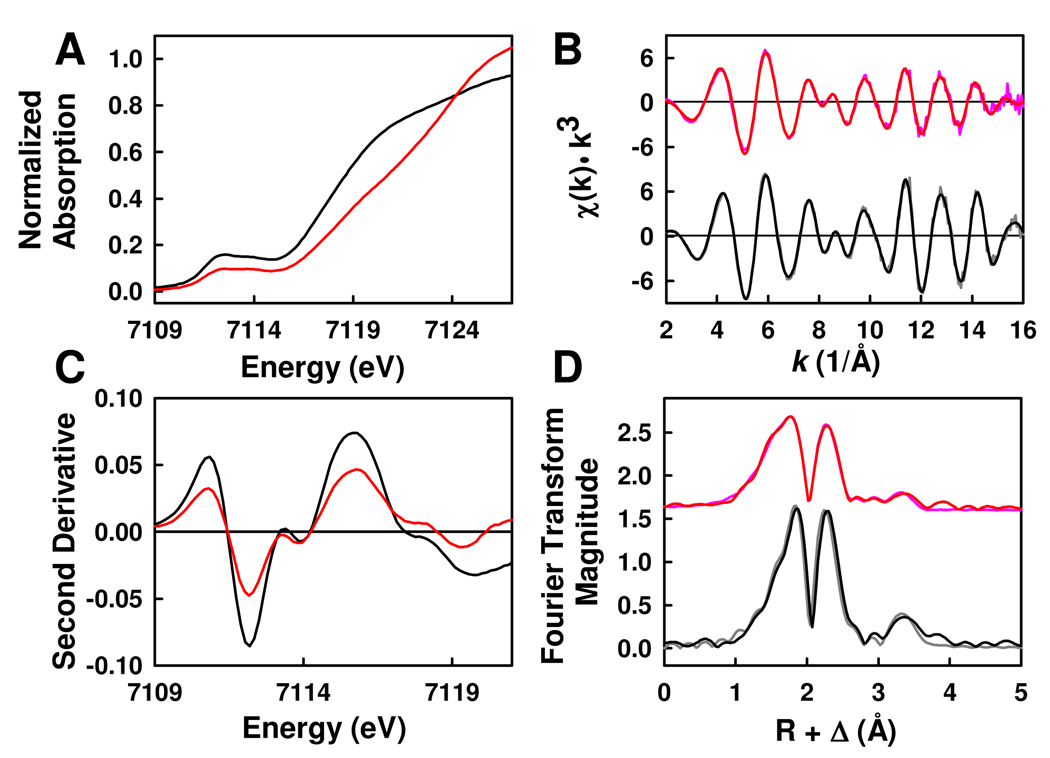
The structural similarities and dissimilarities of NMF-extracted FeVco and FeMoco are further explored by Fe K-edge XAS/EXAFS analyses. The XAS spectra of both isolated cofactors contain a distinct pre-edge component at approximately 7110 eV (Figure 4A). The pre-edge feature of FeVco is of noticeably less intensity than that of FeMoco, suggesting that, on average, the iron coordination in FeVco is less distorted from centro-symmetry. Nevertheless, the two pre-edge features bear a strong resemblance to each other, indicating that the two cofactors do share some degree of similarity in oxidation state and coordination geometry.12 Moreover, the pre-edges of both cofactors can be resolved into two constituent peaks that center at 7112.3 and 7113.9 eV, respectively, and the intensity ratio between the two component peaks are nearly identical for both cofactors (Figure 4B). Such a two-component pre-edge was also observed when the XAS spectrum of the protein-bound FeMoco was derived from the P-cluster-subtracted MoFe protein spectrum, suggesting that the “Fe cages” of both isolated cofactors are similar to that of the protein-bound FeMoco.13,14 On the other hand, the rising edges of the isolated cofactors are quite different in shape and energy (Figure 4A). The rising edge of the FeVco is shallower in gradient and less distinctive in shape compared to that of the FeMoco, and its inflection point around 7117 eV is shifted by ~1 eV to higher energy (Figure 4A). These observations are strongly indicative of a difference in the environment of the Fe atoms in the two isolated cofactors.14,15
Figure 4.
(A) Fe K-edge XAS spectra and (B) smoothed second derivatives for NMF-extracted FeVco (red) and FeMoco (black). (C) Fe K-edge EXAFS and (D) Fourier transforms of data (pink) and fits (red) for FeVco, and those of data (gray) and fits (black) for FeMoco. Data have been normalized to one iron absorber.
The basic fits to the EXAFS of FeVco and FeMoco, which involve the Fe/S backscattering shells in the two cofactors, are roughly equivalent. However, FeMoco is best fit with 3 S atoms at 2.22 Å and 3 Fe atoms at 2.64 Å; whereas FeVco is best fit with 2.5 S atoms at 2.23 Å and 2.5 Fe atoms at 2.63 Å (Table 2). Thus, although the ratio of Fe:S coordination is 1:1 for both cofactors, the overall Fe/S coordination in FeVco is clearly less than that in FeMoco. Such a decrease in Fe/S coordination is “compensated” by an increase in the light atom coordination of the Fe atoms in FeVco, as inclusion of 1.5 O atoms at 1.97 Å and 1.5 C atoms at 3.07 Å results in a significant improvement of the fits to the FeVco data (Table 2). The presence of an increased amount of the light O atoms near the Fe atoms of FeVco is consistent with the reduction in pre-edge intensity and the shift of the rising edge to higher energy (Figure 4A). Moreover, the distance of C atoms is typical of O-bound Fe atoms that are ligated by NMF or other carboxylic acid derivatives.16 Contrary to the FeVco fits, the FeMoco fits are enhanced by including 0.5 O atom at 1.96 Å and no C atom at all (Table 2), suggesting that FeMoco does not interact as strongly as FeVco does with NMF in the isolated state.
Table 2.
Final fits for the Fe K-edge EXAFS data over the k-range of 2–16 Å−1. Coordination number (N), interatomic distance (R, Å), mean-square thermal and static disorder in distance (σ2, Å2), and EXAFS threshold energy adjustment from 7030 eV (ΔE0, eV) were varied in the fits. Estimated errors are ± 0.02 Å in R, ± 0.0001 Å2 in σ2, and ± 20% in N. The goodness of fit, F, is defined as F = [Σk6(χexper. − χcalc.)2/Σk6(χexper.)2]0.5.
| FeMoco | FeVco | |||||
|---|---|---|---|---|---|---|
| No. | R (Å) | Σ2 (Å2) | No. | R (Å) | σ2 (Å2) | |
| Scatterer | ||||||
| Fe-S | 3 | 2.24 | 0.0038 | 2.5 | 2.23 | 0.0049 |
| Fe-Fe short | 3 | 2.64 | 0.0042 | 2.5 | 2.63 | 0.0061 |
| Fe-O | 0.5 | 1.96 | 0.0048 | 1.5 | 1.97 | 0.0067 |
| Fe-C | - | - | - | 1.5 | 3.07 | 0.0015 |
| Fe-Mo | 0.5 | 2.68 | 0.0016 | - | - | - |
| Fe-Fe | 0.5 | 2.87 | 0.0028 | - | - | - |
| Fe-(Fe/V) | - | - | - | 1 | 2.88 | 0.0050 |
| Fe-Fe long | 1 | 3.70 | 0.0022 | 1 | 3.69 | 0.0051 |
| ΔE0 (eV) | −10.6 | −13.5 | ||||
| Weighted F | 0.121 | 0.155 | ||||
Despite their differences in light atom coordination, FeVco and FeMoco have, in common, a long-range Fe backscatterer at ~3.70 Å in their EXAFS fits (Table 2). This Fe atom, which belongs to the opposite cubane of the absorbing Fe atom, generates a cross-cluster signal that is not only representative of a long-range order of the cluster, but also suggestive of the integrity of the cofactors following extraction.17–19 In addition, both cofactors contain intermediate-range metal scatterers in the EXAFS fits. FeMoco is best fit by placing 0.5 Mo atom at 2.68 Å and 0.5 Fe atom at 2.87 Å, which reflects an equal contribution of the two terminal metal atoms (i.e., Mo and the opposite Fe) to the average Fe environment (Table 2); whereas FeVco is equally well fit by either an Fe or a V backscatterer, resulting in a distance of 2.88 Å (Table 2). Attempts to split this shell into two components (i.e., 0.5 Fe and 0.5 V) were not successful. It is worth noting that only a modest gain in fit quality is obtained by including two distinct terminal atom backscattering paths in the fits to the FeMoco data. Nevertheless, thus-calculated distances are consistent with those in the FeMoco structures previously derived from EXAFS or X-ray crystallographic studies.2,13
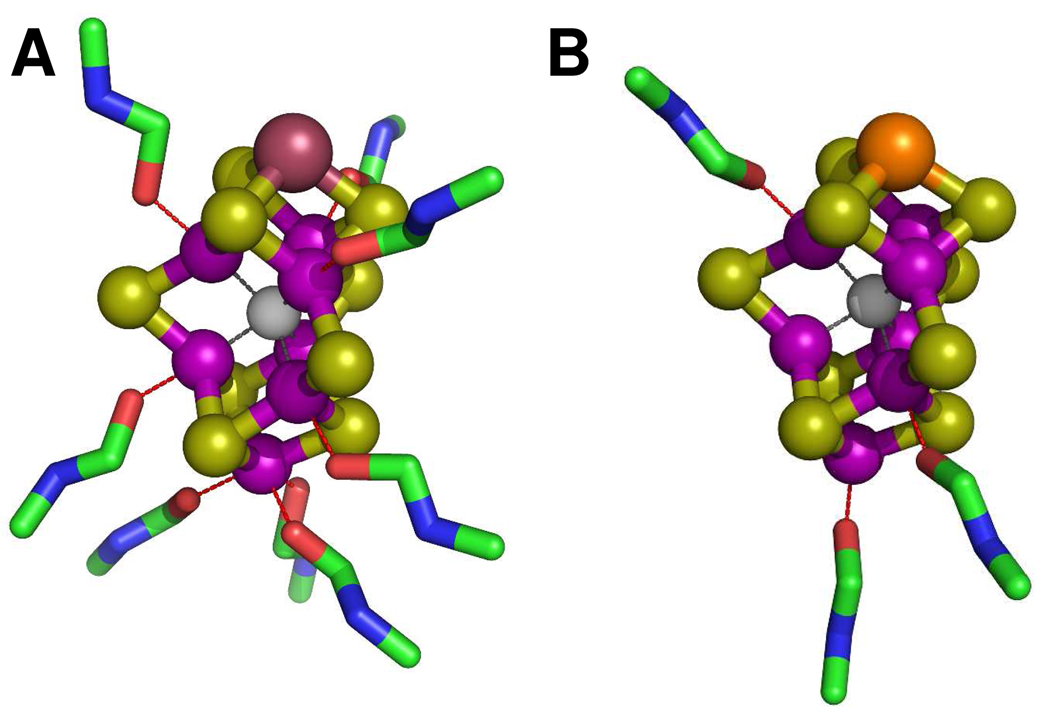
Structural models can be proposed for both isolated cofactors on the basis of their EXAFS fits, (Figure 5). Given the significant similarities of the two cofactors in coordination type, number and distance, it is not surprising that FeVco closely resembles FeMoco in topology. However, contrary to FeMoco, FeVco is coordinated by a well-defined sphere of NMF molecules in the isolated state, suggesting that the electronic structure of FeVco is distinct from that of FeMoco in the absence of a coordinating protein pocket. Combined observations from both the intensities of the pre-edges and the best fits to the EXAFS spectra suggest that, compared to the tetrahedral/trigonal pyramidal structure of the isolated FeMoco, isolated FeVco likely adopts a conformation that is of more octahedral symmetry (Figure 5). It is plausible that the unique electronic properties of the isolated FeVco render it more elongated in structure and more susceptible to solvent binding.20 This argument would be consistent with the observation that the average of the terminal metal backscattering paths in FeVco (i.e., 1 Fe/V at 2.88 Å) is longer than the paths in FeMoco (i.e., 0.5 Mo at 2.68 Å and 0.5 Fe at 2.87 Å) (Figure 5). It should be mentioned that, while many viable models exist for the isolated cofactors, those proposed herein are fully supported by the XAS/EXAFS data and, thus, highly probable from a chemical perspective.
Figure 5.
Structural models of isolated FeVco (A) and FeMoco (B) in NMF. The structural models were adapted from the crystallographic coordinates of the MoFe protein2 but modified for distances on the basis of the EXAFS fits. The atoms are colored as follows: V, plum, Mo, orange; Fe, purple; S, lime; X (C, N or O), light gray; O, red; C, green; N, blue. The models reflect the EXAFS-derived coordination numbers of solvent molecules rather than the actual binding sites of these molecules.
Conclusion
Combined results from metal, activity and spectroscopic analyses provide strong evidence that we have successfully isolated FeVco from A. vinelandii. Compared to the earlier FeVco extract from A. chroococum,5 FeVco isolated herein exhibits a much stronger signal in EPR spectroscopy and significantly improved activities in reconstitution analyses, thereby permitting an unambiguous interpretation of the electronic/catalytic properties of the isolated cofactor. Moreover, contrary to the previous report,5 our FeVco preparation not only carries a characteristic ability to reduce C2H2 to C2H6, but also possesses the capability to reduce N2 to NH3, suggesting that FeVco has been extracted as a complete entity. The improvement in FeVco extraction not only facilitates the proposal of the first EXAFS-based structural model of the isolated FeVco, but also lays the foundation for future catalytic and structural investigations of this nitrogenase cofactor. V K-edge XAS analysis of the isolated FeVco will be pursued in the future, which will permit a further refinement of the structural model and a better understanding of the physiochemical properties of this unique cluster.
3. Experimental Procedures
Cell Growth, Protein Purification and Isolation of Cofactors
A. vinelandii strains YM68A (expressing His-tagged VFe protein), YM13A (expressing His-tagged MoFe protein) and DJ1143 (expressing His-tagged ΔnifB MoFe protein) were grown in 180 L batches in a 200 L New Brunswick fermentor (New Brunswick Scientific, Edison, NJ) in Burke’s minimal medium supplemented with 2 mM ammonium acetate. In the case of YM68A, the molybdenum in Burke’s medium was replaced by an equal amount of vanadium. The growth rate was measured by cell density at 436 nm using a Spectronic 20 Genesys (Spectronic Instruments, Westbury, NY, USA). Cells were harvested in the late exponential phase using a flow-through centrifugal harvester (Cepa, Lahr, Germany), and the cell paste was washed with a buffer containing 50 mM Tris–HCl (pH 8.0). Published methods were used for the purification of His-tagged VFe and MoFe proteins4,21–23 and non-tagged nifH- and vnfH-encoded Fe proteins.24 FeVco and FeMoco were extracted by the acid treatment method into N-methylformamide (NMF) from 1.5 g VFe and MoFe protein, respectively, as described in detail previously.10
Activity Assays and Metal Analysis
Vanadium,25 iron,26 molybdenum,27 and acid labile sulfide28,29 were determined by published methods. Maximum activation of the FeMoco-deficient ΔnifB MoFe protein by NMF-extracted FeVco or FeMoco in N-methylformamide were determined as previously described.24 The products H2, C2H4 and C2H6 were analyzed as published elsewhere,30 and ammonium was determined by a high-performance liquid chromatography fluorescence method.31
Electron Paramagnetic Resonance Spectroscopy
All electron paramagnetic resonance spectroscopy (EPR) samples were prepared in a Vacuum Atmospheres dry box (Hawthorne, CA, USA) at an oxygen level of less than 4 ppm. All NMF-extracted cofactor samples contained 2 mM dithionite (Na2S2O4), and some of them contained either 10 mM thiophenol or 10 mM 1,4-benzene dithiol in addition. The reconstituted MoFe protein samples were prepared by incubating 12.5 mg ΔnifB MoFe protein with 60 nmol NMF-extracted FeVco or FeMoco for 5 minutes at 30°C in 2.5 ml buffer containing 25 mM Tris–HCl (pH 8.0), 10% glycerol and 2 mM Na2S2O4. Subsequently, the reconstituted protein samples were concentrated and the excess cofactor was removed by gel filtration chromatography (Sephadex G-25, GE Healthcare). All protein samples were adjusted to a final concentration of 30 mg/ml and contained 10% glycerol, 2 mM Na2S2O4 and 25 mM Tris–HCl (pH 8.0). Protein samples containing FeMoco (wild-type MoFe protein or FeMoco-reconstituted MoFe protein) have approximately 0.3 mM Mo; whereas protein samples containing FeVco (wild-type VFe protein or FeVco-reconstituted MoFe protein) have approximately 0.25 mM V. Isolated FeMoco and FeVco samples contain 1.5 mM Mo and 1.5 mM V, respectively. Volume-calibrated, clear fused-quartz EPR tubes (Wilmad-LabGlass, Vineland, NJ, USA) were used for EPR experiments. Spectra were collected in perpendicular mode using a Bruker ESP 300 Ez spectrometer (Bruker, Billerica, MA, USA) interfaced with an Oxford Instruments ESR-9002 liquid helium continuous-flow cryostat (Oxford Instruments, Oxford, UK). All spectra were recorded using a gain of 5 × 104, a modulation frequency of 100 kHz, a modulation amplitude of 5 G, a microwave frequency of 9.62 GHz, and a power of 50 mW. Temperatures at which the spectra were taken are indicated in the figure legends.
XAS Data Collection
XAS K-edge data were collected at SSRL beam line 9-3, a 2.0 Tesla 16-pole wiggler side station specifically designed for dilute XAS experiments. SPEAR3 storage ring conditions during data acquisition were 3 GeV and 80 to 100 mA. Incident x-ray photon energy was tuned using a double-crystal Si(220) monochromator. A flat, bent pre-monochromator rhodium-coated mirror was employed for rejection of higher order harmonics and vertical collimation, while a second post-monochromator toroidal rhodium-coated mirror was used for beam focusing. XAS data were collected on NMF-extracted FeMoco and FeVco samples, which contained 2 mM Na2S2O4 and 11.7 mM and 11.1 mM Fe, respectively. These samples were loaded in 1 mm Lucite cells with Kapton tape windows (holding ~95 µl sample) and flash frozen in liquid nitrogen/pentane slush. Samples were maintained at a constant 10 K during data collection by an Oxford Instruments CF1208 continuous flow liquid-He cryostat. EXAFS spectra were collected via detection of Fe Kα fluorescence by a Canberra 30-element solid-state Ge detector with Soller slits and a Mn filter placed between the detector and cryostat to attenuate elastic/inelastic scattering and Fe Kβ fluorescence signal. An iron-foil standard spectrum, measured in tandem with the protein sample scans, was used to calibrate the Fe K-edge x-ray energy to a first inflection point of 7111.2 eV. A total of 12 and 9 scans were collected on the isolated FeVco and FeMoco samples.
XAS Data Analysis
After elimination of defective detector channels and aberrant scans, an average file was generated from the remaining scans using EXAFSPAK.32 The program PYSPLINE was used to subtract a control point delimited first order background from the entire data set, then to generate a spline function to model background absorption, through the EXAFS region.33 For FeMoco, a four-region spline was chosen with 2, 3, 3 order polynomials over the post edge region, while for FeVco best results were obtained with a five-region spline function of the orders 2, 3, 3, 3. The data were normalized to have an edge jump of 1.0 at 7130 eV. The EXAFS spectra were fit over the k-range of 2–16 Å−1 for both samples using the least squares fitting program OPT.32 Ab initio theoretical amplitude and phase functions were generated by FEFF 7.0 using distances derived from the 1M1N crystallographic model.2,34 This model was iteratively refined throughout the fitting process as needed to generate progressively better fits to the EXAFS spectra. Throughout fitting, the amplitude reduction factor was held at 1.0. Consistent with previously described methods, both the interatomic distance (R) between the absorber and backscatter and the mean-square thermal and static deviation (σ2) were allowed to vary for all fit component paths.13,35,36 The threshold energy (ΔE0) was allowed to float, but constrained to be the same for all paths. Coordination numbers (N) were adjusted logically to generate chemically viable fits with the greatest degree of agreement with the EXAFS data and its corresponding Fourier transform. Numerous paths were systematically included and excluded to fully probe the average atomic environment at iron.
Acknowledgment
This work was supported by Herman Frasch Foundation Grant 617-HF07 (M.W.R.), NIH Grant GM 67626 (M.W.R.) and NIH Grant RR 01209 (K.O.H.). SSRL operations are funded by the Department of Energy, Office of Basic Energy Science, and the SSRL Structural Molecular Biology Program by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the DOE Office of Biological and Environmental Research.
REFERENCES
- 1.Burgess BK, Lowe DJ. Chem. Rev. 1996;96:2983–3012. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 2.Einsle O, Tezcan FA, Andrade SLA, Schmid B, Yoshida M, Howard JB, Rees DC. Science. 2002;297:1696–1700. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- 3.Eady RR. Chem. Rev. 1996;96:3013–3030. doi: 10.1021/cr950057h. [DOI] [PubMed] [Google Scholar]
- 4.Lee CC, Hu Y, Ribbe MW. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9209–9214. doi: 10.1073/pnas.0904408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith BE, Eady RR, Lowe DJ, Gormal C. Biochem. J. 1988;250:299–302. doi: 10.1042/bj2500299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilboa-Garber N. Anal. Biochem. 1971;43:129–133. doi: 10.1016/0003-2697(71)90116-3. [DOI] [PubMed] [Google Scholar]
- 7.Miller RW. Anal. Biochem. 1970;35:181–192. doi: 10.1016/0003-2697(70)90024-2. [DOI] [PubMed] [Google Scholar]
- 8.Siegel LM. Anal. Biochem. 1965;11:126–132. doi: 10.1016/0003-2697(65)90051-5. [DOI] [PubMed] [Google Scholar]
- 9.Suhara K, Takemori S, Katagiri M, Wada K, Kobayashi H, Matsubara H. Anal. Biochem. 1975;68:632–636. doi: 10.1016/0003-2697(75)90659-4. [DOI] [PubMed] [Google Scholar]
- 10.Burgess BK. Chem. Rev. 1990;90:1377–1406. [Google Scholar]
- 11.Rawlings J, Shah VK, Chisnell JR, Brill WJ, Zimmerman R, Münck E, Orme-Johnson WH. J. Biol. Chem. 1978;253:1001–1004. [PubMed] [Google Scholar]
- 12.Sarangi R, Aboelella N, Fujusawa K, Tolman WB, Hedman B, Hodgson KO, Solomon EI. J. Am. Chem. Soc. 2006;128:8286–8296. doi: 10.1021/ja0615223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corbett MC, Hu Y, Fay AW, Ribbe MW, Hedman B, Hodgson KO. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1238–1243. doi: 10.1073/pnas.0507853103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westre TE, Kennepohl P, DeWitt JG, Hedman B, Hodgson KO, Solomon EI. J. Am. Chem. Soc. 1997;119:6297–6314. [Google Scholar]
- 15.Randall CR, Shu L, Chiou Y-M, Hagen KS, Ito M, Kitajima N, Lachicotte RJ, Zang Y, Que LJ. Inorg. Chem. 1995;34:1036–1039. [Google Scholar]
- 16.Stremple P, Baenziger NC, Coucouvanis D. J. Am. Chem. Soc. 1981;103:4601–4603. [Google Scholar]
- 17.Harvey I, Arber JM, Eady RR, Smith BE, Garner CD, Hasnain SS. Biochem. J. 1990;266:929–931. [PMC free article] [PubMed] [Google Scholar]
- 18.Arber JM, Flood AC, Garner CD, Gormal CA, Hasnain SS, Smith BE. Biochem. J. 1988;252:421–425. doi: 10.1042/bj2520421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christiansen J, Tittsworth RC, Hales BJ, Cramer SP. J. Am. Chem. Soc. 1995;117:10017–10024. [Google Scholar]
- 20.Lovell T, Torres RA, Han W-G, Liu T, Case DA, Noodleman L. Inorg. Chem. 2002;41:5744–5753. doi: 10.1021/ic020474u. [DOI] [PubMed] [Google Scholar]
- 21.Hu Y, Fay AW, Schmid B, Makar B, Ribbe MW. J. Biol. Chem. 2006;281:30534–30541. doi: 10.1074/jbc.M605527200. [DOI] [PubMed] [Google Scholar]
- 22.Ribbe MW, Hu Y, Guo M, Schmid B, Burgess BK. J. Biol. Chem. 2002;277:23469–23476. doi: 10.1074/jbc.M202061200. [DOI] [PubMed] [Google Scholar]
- 23.Hu Y, Fay AW, Dos Santos PC, Naderi F, Ribbe MW. J. Biol. Chem. 2004;279:54963–54971. doi: 10.1074/jbc.M408983200. [DOI] [PubMed] [Google Scholar]
- 24.Burgess BK, Jacobs DB, Stiefel EI. Biochim. Biophys. Acta. 1980;614:196–209. doi: 10.1016/0005-2744(80)90180-1. [DOI] [PubMed] [Google Scholar]
- 25.Fishman MJ, Skougstad MW. Anal. Chem. 1964;36:1643–1647. [Google Scholar]
- 26.Van de Bogart M, Beinert H. Anal. Biochem. 1967;20:325–334. doi: 10.1016/0003-2697(67)90038-3. [DOI] [PubMed] [Google Scholar]
- 27.Clark LJ, Axley JH. Anal. Biochem. 1955;27:2000–2003. [Google Scholar]
- 28.Tonsager SR, Averill BA. Anal. Biochem. 1980;102:13–15. doi: 10.1016/0003-2697(80)90309-7. [DOI] [PubMed] [Google Scholar]
- 29.Chen J-S, Mortenson LE. Anal. Biochem. 1977;79:157–165. doi: 10.1016/0003-2697(77)90390-6. [DOI] [PubMed] [Google Scholar]
- 30.Gavini N, Burgess BK. J. Biol. Chem. 1992;267:21179–21186. [PubMed] [Google Scholar]
- 31.Corbin JL. Appl. Environ. Microbiol. 1984;47:1027–1030. doi: 10.1128/aem.47.5.1027-1030.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.George GN. EXAFSPAK. Stanford, CA: Stanford Synchrotron Radiation Laboratory; 1990. [Google Scholar]
- 33.Tenderholt A. PYSPLINE. Stanford, CA: Stanford Synchrontron Radiation Laboratory; 2006. [Google Scholar]
- 34.Rehr JJ, Albers RC. Rev. Mod. Phys. 2000;72:621–654. [Google Scholar]
- 35.Hu Y, Corbett MC, Fay AW, Webber JA, Hodgson KO, Hedman B, Ribbe MW. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17119–17124. doi: 10.1073/pnas.0602647103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickering IJ, George GN, Dameron CT, Kurz B, Winge DR, Dance IG. J. Am. Chem. Soc. 1993;115:9498–9505. [Google Scholar]