TABLE 3.
QTL detected for DGY, GM, and SD in F3 and LHRF-F3 populations
| Chr | Conventional F3 |
LHRF-F3 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Left marker | Pos (cM) | CI (cM) | LOD | R2 | Add | Left marker | Pos (IcM) | CI (IcM) | LOD | R2 | Add | |
| DGY (ton ha−1) | ||||||||||||
| 1 | bnlg1083 | 70 | 56–84 | 6.53 | 4.44 | −1.411 | Umc1397 | 226 | 218–240 | 4.23 | 3.51 | −0.927 |
| 1 | umc161 | 254 | 242–272 | 5.12 | 2.77 | 1.250 | Umc161 | 728 | 716–754 | 5.4 | 5.11 | 1.021 |
| 2 | gsy53b | 88 | 40–168 | 3.43 | 2.25 | 0.882 | ||||||
| 2 | Bnlg2248 | 208 | 190–220 | 3.81 | 2.73 | −0.900 | ||||||
| 2 | csu143 | 112 | 108–122 | 16.05 | 9.52 | −2.327 | ||||||
| 2 | phi127 | 462 | 448–476 | 6.69 | 8.43 | −1.291 | ||||||
| 2 | umc137 | 184 | 156–204 | 5.14 | 2.73 | −1.381 | ||||||
| 4 | Umc1511 | 200 | 174–226 | 6.08 | 6.04 | −1.232 | ||||||
| 4 | umc1101 | 212 | 186–230 | 4.16 | 5.88 | −1.131 | csu9b | 476 | 448–506 | 4.46 | 3.20 | −1.145 |
| 4 | Umc1532 | 550 | 538–564 | 3.89 | 2.98 | −0.868 | ||||||
| 5 | Umc83b | 314 | 296–324 | 3.43 | 3.59 | −0.888 | ||||||
| 6 | Bnlg249 | 126 | 108–138 | 6.59 | 9.10 | −1.200 | ||||||
| 7 | umc116 | 68 | 52–106 | 5.24 | 2.72 | 1.332 | ||||||
| 8 | umc152 | 62 | 50–78 | 4.74 | 3.38 | 1.347 | ||||||
| 8 | umc89 | 100 | 76–120 | 3.02 | 2.94 | 1.105 | ||||||
| 9 | csu147 | 204 | 190–236 | 3.76 | 2.60 | −0.939 | ||||||
| 10 | umc1196 | 128 | 118–134 | 7.04 | 5.91 | 1.382 | ||||||
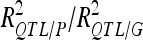 |
48.4 | /59.75 | 41.5 | /56.85 | ||||||||
| GM (%) | ||||||||||||
| 2 | umc134l | 102 | 94–112 | 6.33 | 4.05 | 0.374 | ||||||
| 2 | umc5a | 126 | 114–144 | 3.2 | 2.55 | −0.244 | ||||||
| 2 | umc137 | 172 | 160–186 | 7.07 | 4.30 | −0.289 | ||||||
| 3 | umc1892 | 22 | 0–72 | 2.93 | 2.00 | 0.21 | bnl13.05b | 176 | 166–188 | 3.67 | 3.22 | −0.252 |
| 3 | umc1767 | 158 | 148–180 | 4.59 | 3.42 | −0.238 | ||||||
| 4 | gsy4 | 68 | 62–76 | 19.66 | 18.02 | −0.51 | Mmc0471 | 154 | 142–188 | 5.98 | 8.94 | −0.308 |
| 5 | umc2115 | 76 | 58–100 | 2.72 | 1.6 | −0.171 | ||||||
| 5 | bnl7.71 | 330 | 314–350 | 3.54 | 4.29 | −0.228 | ||||||
| 6 | bnlg1702 | 130 | 118–144 | 4.59 | 2.92 | −0.213 | ||||||
| 8 | Bnlg1031 | 370 | 330–392 | 3.77 | 2.69 | −0.254 | ||||||
| 9 | phi027 | 56 | 46–66 | 6.62 | 5.23 | −0.280 | ||||||
| 9 | bnl5.09 | 378 | 370–386 | 3.66 | 4.67 | −0.230 | ||||||
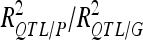 |
45.5 | /55.49 | 20.6 | /24.82 | ||||||||
| SD (days) | ||||||||||||
| 1 | umc1166 | 48 | 28–62 | 6.49 | 5.58 | 0.391 | ||||||
| 1 | umc84 | 282 | 268–292 | 6.45 | 4.76 | 0.366 | Bnlg2123 | 814 | 800–836 | 3.33 | 5.18 | 0.287 |
| 3 | umc1892 | 22 | 6–46 | 3.71 | 2.82 | 0.328 | ||||||
| 3 | dupssr23 | 270 | 240–286 | 3.36 | 3.43 | 0.307 | ||||||
| 5 | mmc0261 | 90 | 82–110 | 4.37 | 3.43 | 0.314 | Umc43 | 250 | 240–260 | 8.25 | 9.63 | 0.559 |
| 5 | Umc1110 | 324 | 312–340 | 4.79 | 5.05 | −0.385 | ||||||
| 6 | gsy189 | 20 | 8–36 | 3.19 | 2.67 | −0.243 | ||||||
| 6 | umc62 | 178 | 156–196 | 2.84 | 2.42 | −0.246 | ||||||
| 8 | umc152 | 64 | 40–86 | 3.51 | 2.88 | 0.324 | ||||||
| 8 | umc89 | 96 | 90–104 | 10.95 | 7.30 | 0.56 | ||||||
| 8 | umc1384 | 180 | 136–190 | 4.53 | 3.59 | −0.300 | ||||||
| 9 | phi027 | 56 | 48–64 | 6.47 | 6.28 | −0.376 | ||||||
| 9 | umc1137 | 200 | 180–208 | 3.55 | 2.60 | 0.261 | Umc1137 | 472 | 456–492 | 4.72 | 5.07 | 0.376 |
| 10 | umc44 | 102 | 86–136 | 5.27 | 3.88 | 0.315 | ||||||
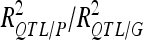 |
45.7 | /54.40 | 26 | /30.23 | ||||||||
For each detected QTL are indicated the chromosome (Chr), the left marker flanking the QTL (left marker), the estimated position (Pos) on the linkage map of the population, the 95% confidence interval of the QTL position (CI), the LOD score, the percentage of phenotypic variance explained by the QTL (R2), and the additive effect (associated with F252 allele). 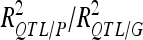 is the proportion of phenotypic variance/genotypic variance explained by all the detected QTL for a given trait. QTL showing significant digenic epistasis (P < 0.01) are underlined. QTL involved in epistatic interaction with two different QTL are double underlined.
is the proportion of phenotypic variance/genotypic variance explained by all the detected QTL for a given trait. QTL showing significant digenic epistasis (P < 0.01) are underlined. QTL involved in epistatic interaction with two different QTL are double underlined.
