Abstract
Adeno-associated viral (AAV) capsid proteins, thought to be a rate-limiting step in the production of recombinant AAV (rAAV), are translated from spliced mRNAs. Improvement of the native AAV nonconsensus donor sequence increases splicing yet leaves the relative levels of VP1- and VP2/3-encoding mRNAs unchanged, and thus provides a means to increase delivery of correct ratios of AAV capsid proteins. This effect is independent of the AAV serotype used, and occurs whether the rep and cap genes supplied in trans are on the same or separate expression vectors. In the split-vector system, replacement of the more traditionally used cytomegalovirus promoter with that of the AAV5 P41 promoter allowed for even greater levels of splicing, and together with an improved intron donor, led to a 10- to 15-fold increase in the levels of splicing, rAAV production, and transduction compared with levels achieved by traditional cotransfection methods. Thus, the enhancement of splicing presents a useful method to enhance rAAV production via transient transfection.
Introduction
Adeno-associated virus (AAV) is a small, nonenveloped, single-stranded DNA (ssDNA) virus that was initially discovered as a contaminant of adenoviral stocks (Atchison et al., 1966; Hoggan et al., 1966; Blacklow et al., 1967). AAV is a member of the genus Dependovirus of the subfamily Parvovirinae and its efficient replication requires coinfection with a helper virus such as adenovirus or herpesvirus (Bowles et al., 2006). In the presence of the AAV Rep protein, wild-type AAV can integrate efficiently into human chromosome 19 (Kotin et al., 1990; Samulski et al., 1991). In the absence of Rep, recombinant AAV (rAAV) genomes have been shown to persist episomally in the nucleus of many cell types (Yan et al., 2006). Because AAV is capable of transducing and persisting in a number of cell types, because it has not been found to be associated with any human disease, and because of its low observed immunogenicity, it has become a useful gene therapy candidate for a number of diseases (Carter, 2006).
A typical method of generating rAAV involves cotransfection of three plasmids: an inverted terminal repeat (ITR)-containing plasmid, which has the rep and cap genes of AAV replaced with that of a promoter and gene of interest to be expressed; an adenoviral (Ad) helper plasmid, which together with host 293 cells provides the minimal gene products required for AAV replication; and finally, the AAV rep and cap genes, supplied in trans from a third vector, for encapsidation. The generation of high levels of rAAV remains a challenge, and production of rAAV to the levels needed for human clinical trials remains costly and labor-intensive. Increased yields of rAAV have been achieved by repressing expression of Rep78 and Rep68 protein levels, either by mutation of their initiating ATG to ACG or by replacing the endogenous P5 promoter with the weaker mouse mammary tumor virus long terminal repeat (MMTV-LTR) promoter; by increasing capsid production through the use of vectors that express the rep and cytomegalovirus (CMV)-driven cap genes individually; and by the use of packaging cell lines or baculoviral expression systems (Li et al., 1997; Vincent et al., 1997; Xiao et al., 1998; Urabe et al., 2000, 2002; Cao et al., 2002; Li and Samulski, 2005; Negrete and Kotin, 2008).
The three capsid proteins of AAV are generated primarily from spliced mRNAs (Becerra et al., 1988; Trempe and Carter, 1988), and their relative abundances determine the levels of the individual capsid proteins that are produced. The 5′ splice sites of all human and nonhuman primate AAVs (with the exception of AAV5) are identical (CAG|GTACCA), and differ from the U1 small nuclear ribonucleoprotein (snRNP) consensus binding site CAG|GTAAGT (differences between these two sequences are italicized). Therefore, the enhancement of splicing of the capsid gene pre-mRNAs by improving the intron donor presents a potentially useful avenue to significantly increase rAAV titers. Such improvements would not be expected to alter the relative ratios of the mRNAs that individually encode VP1, VP2, and VP3, which are produced by alternative use of the intron acceptors.
In this report, we describe a simple mechanism to increase the levels of rAAV production during transient transfection. By modifying the splice donor site of the small internal AAV intron to resemble more closely the sequence of the consensus U1 snRNP-binding site, we were able to significantly enhance the overall levels of splicing of the capsid-generating P40 pre-mRNA transcripts. Increases in splicing via donor alteration resulted in a parallel increase in both capsid-encoding mRNAs and so did not disrupt the delicate balance between the three capsid proteins. Subsequent increases in rAAV vector production were thus directly proportional to the levels of splicing. We demonstrate that this strategy can be used either in a single-vector system, in which the rep and cap genes are on the same vector, or more robustly, in the dual-vector system that uses the two major AAV open reading frames on separate vectors. In addition, the AAV5 P41 promoter increases both expression and splicing of capsid gene RNAs compared with the cytomegalovirus immediate-early promoter, and together with an improved intron donor, results in levels of splicing, rAAV production, and transduction that were further enhanced as much as 10- to 15-fold compared with levels achieved by current cotransfection methods.
Materials and Methods
Cell lines, AAV packaging plasmids, and viruses
293T cells were propagated as previously described (Mouw and Pintel, 2000). The AAV2 RepCap plasmid, which lacks both ITRs, was described previously (Qiu and Pintel, 2002). The AAV1 RepCap plasmid was generated by inserting bases 181 to 4566 from AAV1 into the EcoRI and XbaI sites of pSK(+) (Stratagene, La Jolla, CA). The AAV6 Rep-Cap plasmid was generated by inserting bases 181 to 4560 from AAV6 into the EcoRI and XbaI sites of pSK(+). The CMV-Cap2 plasmid was previously described (Qiu and Pintel, 2002). “Better” or consensus donor mutations were generated by overlapping polymerase chain reaction (PCR) mutagenesis of the native AAV donor site (CAG|GTACCA) to that of the “better” donor site (CAG|GTACGT) or consensus donor site (CAG|GTAAGT). The AAV2/8 RepCap and CMV-Cap8 plasmids were generous gifts from D. Miller (Fred Hutchinson Cancer Research Center, Seattle, WA) and J. Wilson (University of Pennsylvania, Philadelphia, PA). P41-Cap2 and P41-Cap8 constructs were generated by replacing the region of AAV2 RepCap or AAV2/8 RepCap from the SfiI site at position 544 to the HindIII site at position 1883, with a PCR-amplified P41 promoter from AAV5 (positions 1681 to 1974), using SfiI for the 5′ end and HindIII for the 3′ end.
To generate recombinant AAV, the AAV2 ITR-containing packaging vector pMU2, which contains the green fluorescent protein (GFP)-encoding gene as a reporter gene as described previously (Reed et al., 2006), was used. To supply the four AAV2 Rep proteins according to the split-vector method, the RepSM vector was used, also as previously described (Qiu and Pintel, 2002). Ad helper functions were supplied by pHelper as previously described (Qiu et al., 2002).
Transfections, RNA isolation, and analysis
For RNA analysis, transfections were conducted according to a modified polyethylenimine (PEI) transfection method as described (Reed et al., 2006). Cells were harvested and RNA was isolated 36-42 hr posttransfection (Qiu and Pintel, 2002). RNase protection assays were performed with 10 μg of total RNA as previously described (Schoborg and Pintel, 1991; Naeger et al., 1992).
RNA was isolated after guanidinium isothiocyanate lysis and cesium chloride gradient centrifugation as previously described (Schoborg and Pintel, 1991; Naeger et al., 1992). The AAV2 RP probe, which spans nucleotides 1767–1958 and allows identification of spliced and unspliced P40 transcripts, was described previously (Mouw and Pintel, 2000). AAV1 and AAV6 homologous RP probes were generated by PCR amplification of region 1781–1975 in AAV1 and region 1766–1960 in AAV6, and subsequent cloning into the EcoRI/BamHI sites in pGEM-3Z (Promega, Madison, WI). CMV-Cap RP probes were generated by PCR amplification with a forward CMV primer and the appropriate serotype-specific reverse RP primer, as described above. P41-Cap RP probes were generated by PCR amplification with a forward AAV5 P41 primer and the appropriate serotype-specific reverse RP primer, as described above.
For rAAV production, cells were plated onto 100-mm dishes the day before transfection, and a total of 20 μg of DNA was transfected per plate according to the PEI method as described (Reed et al., 2006). For triple transfections, reactions were carried out at a ratio of 1:1:2 (pMU2:Rep-Cap:pHelper). For quadruple transfections, reactions were carried out at a ratio of 1:1:1:2 (pMU2:Rep:Cap:pHelper). Recombinant virus was harvested 60 hr posttransfection.
Recombinant AAV isolation/quantitative PCR
Sixty hours posttransfection, cells were collected and processed as described (Mayginnes et al., 2006). Briefly, cell pellets were resuspended in 50 mM Tris–1 mM EDTA buffer and subjected to three cycles of freeze–thaw lysis before brief sonication. Lysates were diluted 1:100 and treated with DNase I to remove unpackaged genomic DNA. During DNase I inactivation, samples were diluted a further 1:15 before quantitative PCR (qPCR) analysis. Quantification of packaged rAAV genomes was calculated with SYBR green reagent (Bio-Rad, Hercules, CA) on a 7500 real-time PCR system (Applied Biosystems, Foster City, CA) as described previously. The forward and reverse GFP primers used for qPCR were also described previously (Mayginnes et al., 2006). rAAV production in the absence of pHelper was negligible (data not shown).
Transduction assay
HeLa cells were plated onto 24-well plates 24 hr before infection. After quantification of packaged rAAV genomes by qPCR, aliquots of the crude lysates were standardized, and serial 10-fold dilutions (beginning with 5 × 107 packaged genomes per well) were used for infection. GFP-positive cells were counted and approximate transducing units (TU) of rAAV were scored as the number of GFP-positive cells relative to the total number of cells. Infectious titers were calculated by dividing the genomic copy number (as assayed by qPCR) by the amount of transducing units (Mayginnes et al., 2006). The ssDNA/TU ratios for all rAAVs were tested in the absence of helper Ad, and ranged from approximately 1400 to 2000; this ratio was lower when Ad was used in these assays.
Results and Discussion
At least one of the rate-limiting steps in the production of recombinant AAV is the production of capsids (Vincent et al., 1997). We found our yields of rAAV1 and rAAV6 vectors to be consistently less than those of rAAV2 (data not shown), and because AAV capsid proteins are generated from spliced capsid gene mRNA, we wondered whether reduced levels of splicing could contribute to the difference in rAAV production for these serotypes. We found that, similar to AAV2, the basal level of expression and splicing of AAV1 and AAV6 pre-mRNAs generated from RepCap constructs in 293 cells is low (data not shown). However, in contrast to AAV2, enhancement of splicing of pre-mRNAs generated from AAV1 (Fig. 1, compare lane 1 with lane 4), and AAV6 (data not shown) RepCap constructs, in the presence of pHelper (supplying E2a, E4orf6, and VA RNA), or adenoviral coinfection (data not shown), remained modest.
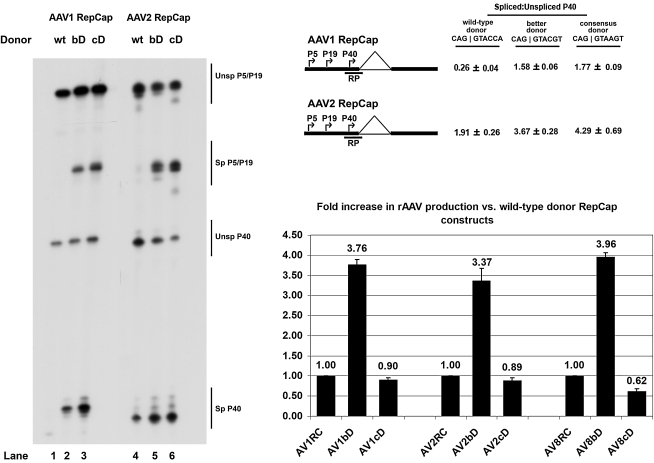
FIG. 1.
Improvement of the nonconsensus AAV donor significantly enhanced overall pre-mRNA splicing, capsid production, and rAAV production. Left: Representative RNase protection assay of AAV1 and AAV2 RepCap plasmids (AV1RC and AV2RC, respectively) with wild-type, “better” (bD), or consensus donor (cD) mutations in the presence of pHelper. Unsp, Unspliced; Sp, Spliced. Top right: Quantification of the relative spliced-to-unspliced ratios of capsid-encoding pre-mRNA. Data are from at least three experiments and show standard deviations. The position of the “RP” RNase protection probe is indicated. Bottom right: Relative levels of rAAV production observed for AAV1, AAV2, and AAV8 with the “better” and consensus donor mutants in relation to levels obtained with the wild-type donors (set to a value of 1.00). Average titers of DNase-resistant rAAV virions per 100-mm dish (with standard deviations in parentheses) were as follows: AV1RC, 1.25 × 1010 (±4.87 × 109); AV1bD, 4.78 × 1010 (±2.16 × 1010); AV1cD, 1.08 × 1010 (±2.75 × 109); AV2RC, 3.43 × 1010 (±1.33 × 1010); AV2bD, 1.15 × 1011 (±4.10 × 1010); AV2cD, 3.04 × 1010 (±1.34 × 1010); AV8RC, 6.97 × 1010 (±2.01 × 1010); AV8bD, 2.78 × 1011 (±9.07 × 1010); AV8cD, 4.45 × 1010 (±1.72 × 1010).
Improvement of nonconsensus AAV donor significantly enhanced overall pre-mRNA splicing, capsid production, and rAAV production
To test the hypothesis that splicing levels of the capsid protein-supplying construct contribute to the levels of production of rAAV, we attempted to increase the levels of spliced mRNAs by generating a series of AAV1 and AAV2 RepCap vectors in which the native, nonconsensus donor sites were improved. We made constructs in which the donors were made fully complementary to the endogenous U1 snRNP interaction site, which also adds a termination codon in the Rep78 and Rep52 open reading frames (ORFs) (C AG|G TAC CA to C AG|G TAA GT, termination signal in boldface, termed the consensus donor [cD]), as well as constructs in which the U1 snRNP-binding donor sequence was improved but did not generate a termination signal (CAG|GTACCA to CAG|GTACGT, introducing a glutamine [CAA] to valine [GTA] substitution, termed the “better” donor [bD]). Wild-type and mutant AAV1 and AAV2 RepCap constructs were then assayed for splicing by RNase protection assays, using homologous probes. As expected, in 293 cells in the presence of pHelper, the overall levels of pre-mRNA splicing of both AAV1 and AAV2 capsid-encoding RNAs were increased significantly when the donors more closely resembled the consensus sequence. For AAV1, the “better” donor and consensus donor improvements led to approximately 6- and 7-fold increases in splicing, respectively (Fig. 1, AV1RepCap bD and cD, compare lane 1 with lanes 2 and 3), whereas for AAV2, the “better” donor and consensus donor improvements led to approximately 2-fold increases (Fig. 1, AV2RepCap bD and cD, compare lane 4 with lanes 4 and 5). A proportional increase in capsid protein production was also detected by Western immunoblotting (data not shown). In addition to an increase in P40 pre-mRNA splicing, we also saw a marked increase in the overall levels of pre-mRNA splicing from the upstream P5 and P19 promoters in both AAV1 and AAV2, which also presumably led to an increase in the relative levels of Rep68 compared with Rep78 and of Rep40 compared with Rep52. That the donor improvements increased splicing of AAV1 RNA to a greater extent than for AAV2 RNA may indicate that splicing of AAV2 RNA is more dependent on the Rep proteins than is splicing of AAV1 RNA.
The “better” and consensus donor mutants were then used as helper plasmids to supply the Rep and capsid proteins needed, together with pHelper, to generate rAAV in 293 cells. The same “better” and consensus donor mutants were also introduced in a hybrid AAV2/AAV8 RepCap vector to examine the effects of increased splicing on the production of rAAV8-encapsidated virions. As shown in the graph in Fig. 1, the use of the “better” donor mutants (which substitute an alanine for a glutamine in the Rep proteins) resulted in a significant increase (approximately 3- to 4-fold) in the numbers of packaged rAAV genomes compared with those generated by the wild-type constructs for all three serotypes. This was likely because of increased relative levels of available AAV capsid proteins; however, increased relative levels of Rep68 and Rep40, compared with Rep78 and Rep52, respectively, may have also played a role. As expected, the consensus donor constructs, which terminate the Rep open reading frame, supported even fewer packaged genomes than the wild-type RepCap constructs, consistent with previous findings suggesting that full-length Rep78 and Rep52 proteins are required for efficient viral production (Chiorini et al., 1998; Jing et al., 2001; Nada and Trempe, 2002; Di Pasquale and Chiorini, 2003; Timpe et al., 2006).
Use of a split Rep/Cap AAV helper system overcame the negative effects of the consensus donor, allowing for even greater levels of rAAV production
AAV helper vectors in which the Rep and capsid proteins are on the same vector risk wild-type AAV contamination originating from low levels of recombination between the RepCap and ITR-containing plasmids. Several strategies have been used to overcome this problem, including the addition of intronic sequences to the RepCap vectors (Allen et al., 1997; Wang et al., 1998; Cao et al., 2000; Li and Samulski, 2005), and the use of a split-vector system in which the rep and cap genes are on different plasmids (Whiteway et al., 2003). We hypothesized that the consensus donor mutants, which introduce a termination codon in the rep gene of Rep-Cap plasmids, may have a greater enhancing effect on vector production in systems in which rep and cap are supplied from separate plasmids.
At present, a common capsid vector of choice in the split-vector system uses the cytomegalovirus immediate-early promoter (CMV-IE) to drive expression of the capsid-encoding pre-mRNA (Whiteway et al., 2003). Interestingly, we have previously shown (Qiu and Pintel, 2002), and show again in this study (Fig. 2, lane 2), that splicing of AAV2 pre-mRNAs expressed from the CMV-IE promoter in 293 cells is relatively inefficient, even in the presence of pHelper. The addition of Rep78 in trans did not further enhance splicing (data not shown). However, substitution of the native nonconsensus AAV2 donor sequence in the CMV-driven cap construct with that of the “better” donor led to an approximate 3- and 4.5-fold increase in the level of splicing in either the absence or presence of pHelper, respectively (Fig. 2, lanes 3 and 5); whereas introduction of the consensus donor led to approximate 10- and 9-fold increases in the levels of splicing in the absence and presence of pHelper, respectively (Fig. 2, lanes 4 and 6). RNase protection assays using a probe across the intron acceptors that allowed for the quantification of the relative usage of the two AAV acceptor sites revealed no significant alteration of the relative levels of A1/A2 splicing (data not shown).
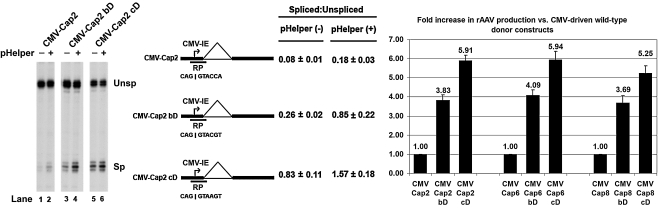
FIG. 2.
Use of a split Rep/Cap AAV helper system overcame the negative effects of the consensus donor, allowing for even greater levels of rAAV production. Left: Representative RNase protection assay of AAV2 CMV-driven capsid-encoding pre-mRNAs in the absence or presence of pHelper. Middle: Quantification of the relative spliced-to-unspliced ratios of capsid-encoding pre-mRNA. Data are from at least three experiments and show standard deviations. The position of the “RP” RNase protection probe is indicated. Right: Relative levels of rAAV production observed in AAV2, AAV6, and AAV8 with the “better” and consensus donor mutants in relation to levels obtained with the wild-type donors (set to a value of 1.00). Average titers of DNase-resistant rAAV virions per 100-mm dish (with standard deviations in parentheses) were as follows: CMV-Cap2, 1.95 × 1010 (±6.75 × 109); CMV-Cap2bD, 7.47 × 1010 (±1.78 × 1010); CMV-Cap2cD, 1.15 × 1011 (±4.82 × 1010); CMV-Cap6, 9.24 × 109 (±6.84 × 108); CMV-Cap6bD, 3.75 × 1010 (±2.41 × 109); CMV-Cap6cD, 5.45 × 1010 (±3.27 × 109); CMV-Cap8, 2.45 × 1010 (±7.27 × 109); CMV-Cap8bD, 8.59 × 1010 (±1.56 × 1010); CMV-Cap8cD, 1.25 × 1011 (±3.06 × 1010).
The “better” and consensus donor mutant AAV2 CMV-Cap constructs were then tested for their abilities to aid in producing rAAV, using this split-vector system. In addition, similar donor mutations were made in AAV6 and AAV8 CMV-driven capsid-encoding constructs. Similarly to the RepCap constructs in Fig. 1, the “better” donor mutant constructs generated 3- to 4-fold increased levels of rAAV production for all three serotypes (Fig. 2). As predicted, the consensus donor mutants increased vector production to even higher levels than the “better” donor mutants (approximately 5- to 6-fold), suggesting that the limitations of using the consensus donor mutants in the RepCap single-vector system were due to truncations in Rep and could be overcome by supplying rep and cap on separate vectors.
The AAV5 P41 promoter allows for more efficient splicing of capsid-encoding AAV pre-mRNA than the CMV promoter, and together with an improved donor, led to the generation of high levels of rAAV
Although the strong CMV promoter is useful for the production of rAAV from a split-vector system, most of the pre-mRNAs generated from these vectors remains unspliced and therefore are poor sources for the capsid proteins. This remains the case even when the donors are improved. We have found that in contrast to the AAV2 P40 promoter, the AAV5 P41 capsid-gene promoter generates a high level of spliced mRNAs in 293 cells because of E1A in the absence of added Rep or additional Ad5 gene products (Qiu et al., 2002; Ye et al., 2006). Therefore, we hypothesized that although the AAV5 P41 promoter does not contain an Rep-binding element (Ye et al., 2006), it could be a useful substitute for the CMV promoter in driving expression of the AAV capsid proteins for rAAV production in 293 cells. This was found to be the case, and more importantly, the levels of spliced pre-mRNA were found to be the predominant species of capsid gene RNA when the nonconsensus AAV2 donor site in these constructs was made consensus (Fig. 3, compare lanes 1–3 with lanes 4 and 5).
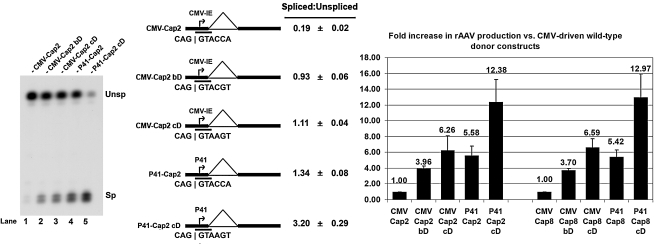
FIG. 3.
The AAV5 P41 promoter allows for more efficient splicing of capsid-encoding AAV pre-mRNA than the CMV promoter, and together with an improved donor, led to the generation of high levels of rAAV. Left: RNase protection assay of CMV- and P41-driven AAV2 capsid-encoding pre-mRNAs in the presence of pHelper. Middle: Quantification of the relative spliced-to-unspliced ratios of capsid-encoding pre-mRNA. Data are from at least three experiments and show standard deviations. The position of the “RP” RNase protection probe is indicated. Right: Relative levels of rAAV production observed in AAV2 and AAV8 with the “better” and consensus donor mutants, in addition to the P41 wild-type and consensus donor mutants, in relation to levels obtained with the CMV-driven wild-type donor capsid-encoding construct (set to a value of 1.00). Titers of rAAVs generated with the CMV-driven wild-type donor construct typically ranged from approximately 9.5 × 109 to 2.0 × 1011 packaged genomes per 100-mm dish. Average titers of DNase-resistant rAAV virions per 100-mm dish (with standard deviations in parentheses) were as follows: CMV-Cap2, 8.59 × 1010 (±3.41 × 1010); CMV-Cap2bD, 3.33 × 1011 (±1.28 × 1011); CMV-Cap2cD, 5.02 × 1011 (±1.72 × 1011); P41-Cap2, 5.05 × 1011 (±2.34 × 1011); P41-Cap2cD, 1.08 × 1012 (±4.40 × 1011); CMV-Cap8, 2.77 × 1011 (±1.57 × 1011); CMV-Cap8bD, 1.10 × 1012 (±6.83 × 1011); CMV-Cap8cD, 2.18 × 1012 (±1.53 × 1012); P41-Cap8, 1.31 × 1012 (±5.89 × 1011); P41-Cap8cD, 4.44 × 1012 (±3.18 × 1012).
These vectors were then tested in the split-vector system to generate rAAV. Consistent with their relatively high basal expression of spliced capsid gene mRNAs, the P41-driven capsid vectors containing the native donor site generated 5- to 6-fold higher levels of rAAV than the CMV-driven capsid constructs containing the wild-type donor; intermediate between CMV-driven constructs bearing the “better” and the consensus donors. The P41-driven Cap constructs containing the consensus donor produced approximately 10- to 15-fold more rAAV than the CMV-driven capsid-gene constructs bearing the wild-type donor, yields approximately twice as high as those of the CMV-driven capsid constructs containing a consensus donor, and significantly higher than any of the RepCap-linked constructs. Similar results were also observed when the P41 promoter was used to drive expression of the AAV8 capsids, again suggesting that this pattern is not serotype specific.
Levels of transduction-capable rAAV correlate well with the increased levels of packaged genomes
Levels of transduction-capable rAAV can be determined by calculating the ratio of packaged ssDNA genomes to transducing units (ssDNA/TU). For rAAV2, this ssDNA/TU ratio is usually between 500 and 1500, as assayed in the absence of Ad (Mayginnes et al., 2006). To determine whether the increase in packaged genomes obtained with the splicing mutants led to a concomitant increase in the numbers of transducing virions, 10-fold serial dilutions of crude cell packaging lysates were used to infect HeLa cells and the number of GFP-positive cells was determined 48 hr postinfection. Recombinant AAV8s generated with the consensus donor mutants, or generated with the AAV5 P41 promoter, were transduced as efficiently, on a vector genome basis, as rAAV8 generated with the CMV-Cap expression vector (Fig. 4).
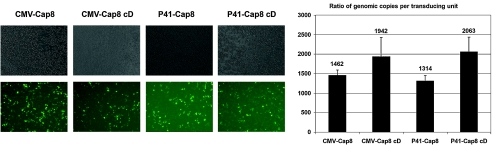
FIG. 4.
Levels of transduction-capable rAAV correlate well with the increased levels of packaged genomes. Shown are the results of a transduction assay of HeLa cells, plated on 24-well dishes 24 hr before infection with 10-fold serial dilutions of rAAV8 (5 × 107 packaged genomes per well). GFP-positive cells were counted 48 hr postinfection. Top left: Phase-contrast microscopy. Bottom left: GFP-positive cells. Right: The number of packaged genomes per well was divided by the approximate number of GFP-positive transduced cells to obtain the efficiency of infection. Values indicated are expressed as a ratio of packaged genomes per transducing unit. The ssDNA/TU ratio for all rAAVs tested in the absence of Ad ranged from approximately 1400 to 2000. Data are from three experiments and show standard deviations.
Thus, improving the levels of spliced capsid gene mRNAs is a useful strategy to improve rAAV production. Why the AAV1 and AAV6 introns remain poorly spliced in the presence of Rep and Ad, even though their donors and acceptors are identical to those of AAV2, is currently under investigation.
Acknowledgments
The authors thank Lisa Burger for outstanding technical assistance; and Gregory Tullis, Jianming Qiu, and Dongsheng Duan for technical assistance, reagents, and advice regarding recombinant AAV production. The authors also thank Dusty Miller and James Wilson for reagents. This study was supported by Public Health Service grants RO1 AI46458 and RO1 AI56310 from the NIAID to D.J.P.
Author Disclosure Statement
No competing financial interests exist.
References
- Allen J.M. Debelak D.J. Reynolds T.C. Miller A.D. Identification and elimination of replication-competent adeno-associated virus (AAV) that can arise by nonhomologous recombination during AAV vector production. J. Virol. 1997;71:6816–6822. doi: 10.1128/jvi.71.9.6816-6822.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchison R.W. Casto B.C. Hammon W.M. Electron microscopy of adenovirus-associated virus (AAV) in cell cultures. Virology. 1966;29:353–357. doi: 10.1016/0042-6822(66)90045-6. [DOI] [PubMed] [Google Scholar]
- Becerra S.P. Koczot F. Fabisch P. Rose J.A. Synthesis of adeno-associated virus structural proteins requires both alternative mRNA splicing and alternative initiations from a single transcript. J. Virol. 1988;62:2745–2754. doi: 10.1128/jvi.62.8.2745-2754.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacklow N.R. Hoggan M.D. Rowe W.P. Isolation of adenovirus-associated viruses from man. Proc. Natl. Acad. Sci. U.S.A. 1967;58:1410–1415. doi: 10.1073/pnas.58.4.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles D.E. Rabinowitz J.E. Samulski R.J. The Genus Dependovirus. Hodder Arnold; London: 2006. [Google Scholar]
- Cao L. Liu Y. During M.J. Xiao W. High-titer, wild-type free recombinant adeno-associated virus vector production using intron-containing helper plasmids. J. Virol. 2000;74:11456–11463. doi: 10.1128/jvi.74.24.11456-11463.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L. During M. Xiao W. Replication competent helper functions for recombinant AAV vector generation. Gene Ther. 2002;9:1199–1206. doi: 10.1038/sj.gt.3301710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter B.J. Clinical Development with Adeno-associated Virus Vectors. Hodder Arnold; London: 2006. [Google Scholar]
- Chiorini J.A. Zimmermann B. Yang L. Smith R.H. Ahearn A. Herberg F. Kotin R.M. Inhibition of PrKX, a novel protein kinase, and the cyclic AMP-dependent protein kinase PKA by the regulatory proteins of adeno-associated virus type 2. Mol. Cell. Biol. 1998;18:5921–5929. doi: 10.1128/mcb.18.10.5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pasquale G. Chiorini J.A. PKA/PrKX activity is a modulator of AAV/adenovirus interaction. EMBO J. 2003;22:1716–1724. doi: 10.1093/emboj/cdg153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggan M.D. Blacklow N.R. Rowe W.P. Studies of small DNA viruses found in various adenovirus preparations: Physical, biological, and immunological characteristics. Proc. Natl. Acad Sci. U.S.A. 1966;55:1467–1474. doi: 10.1073/pnas.55.6.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing X.J. Kalman-Maltese V. Cao X. Yang Q. Trempe J.P. Inhibition of adenovirus cytotoxicity, replication, and E2a gene expression by adeno-associated virus. Virology. 2001;291:140–151. doi: 10.1006/viro.2001.1192. [DOI] [PubMed] [Google Scholar]
- Kotin R.M. Siniscalco M. Samulski R.J. Zhu X.D. Hunter L. Laughlin C.A. McLaughlin S. Muzyczka N. Rocchi M. Berns K.I. Site-specific integration by adeno-associated virus. Proc. Natl. Acad Sci. U.S.A. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. Samulski R.J. Serotype-specific replicating AAV helper constructs increase recombinant AAV type 2 vector production. Virology. 2005;335:10–21. doi: 10.1016/j.virol.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Li J. Samulski R.J. Xiao X. Role for highly regulated rep gene expression in adeno-associated virus vector production. J. Virol. 1997;71:5236–5243. doi: 10.1128/jvi.71.7.5236-5243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayginnes J.P. Reed S.E. Berg H.G. Staley E.M. Pintel D.J. Tullis G.E. Quantitation of encapsidated recombinant adeno-associated virus DNA in crude cell lysates and tissue culture medium by quantitative, real-time PCR. J. Virol. Methods. 2006;137:193–204. doi: 10.1016/j.jviromet.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Mouw M.B. Pintel D.J. Adeno-associated virus RNAs appear in a temporal order and their splicing is stimulated during coinfection with adenovirus. J. Virol. 2000;74:9878–9888. doi: 10.1128/jvi.74.21.9878-9888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nada S. Trempe J.P. Characterization of adeno-associated virus Rep protein inhibition of adenovirus E2a gene expression. Virology. 2002;293:345–355. doi: 10.1006/viro.2001.1286. [DOI] [PubMed] [Google Scholar]
- Naeger L.K. Schoborg R.V. Zhao Q. Tullis G.E. Pintel D.J. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 1992;6:1107–1119. doi: 10.1101/gad.6.6.1107. [DOI] [PubMed] [Google Scholar]
- Negrete A. Kotin R.M. Strategies for manufacturing recombinant adeno-associated virus vectors for gene therapy applications exploiting baculovirus technology. Brief. Funct. Genomic. Proteomic. 2008;7:303–311. doi: 10.1093/bfgp/eln034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J. Pintel D.J. The adeno-associated virus type 2 Rep protein regulates RNA processing via interaction with the transcription template. Mol. Cell. Biol. 2002;22:3639–3652. doi: 10.1128/MCB.22.11.3639-3652.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J. Nayak R. Tullis G.E. Pintel D.J. Characterization of the transcription profile of adeno-associated virus type 5 reveals a number of unique features compared with previously characterized adeno-associated viruses. J. Virol. 2002;76:12435–12447. doi: 10.1128/JVI.76.24.12435-12447.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S.E. Staley E.M. Mayginnes J.P. Pintel D.J. Tullis G.E. Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J. Virol. Methods. 2006;138:85–98. doi: 10.1016/j.jviromet.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Samulski R.J. Zhu X. Xiao X. Brook J.D. Housman D.E. Epstein N. Hunter L.A. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoborg R.V. Pintel D.J. Accumulation of MVM gene products is differentially regulated by transcription initiation, RNA processing and protein stability. Virology. 1991;181:22–34. doi: 10.1016/0042-6822(91)90466-o. [DOI] [PubMed] [Google Scholar]
- Timpe J.M. Verrill K.C. Trempe J.P. Effects of adeno-associated virus on adenovirus replication and gene expression during coinfection. J. Virol. 2006;80:7807–7815. doi: 10.1128/JVI.00198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe J.P. Carter B.J. Alternate mRNA splicing is required for synthesis of adeno-associated virus VP1 capsid protein. J. Virol. 1988;62:3356–3363. doi: 10.1128/jvi.62.9.3356-3363.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urabe M. Shimazaki K. Saga Y. Okada T. Kume A. Tobita K. Ozawa K. Self-amplification system for recombinant adeno-associated virus production. Biochem. Bio-phys. Res. Commun. 2000;276:559–563. doi: 10.1006/bbrc.2000.3521. [DOI] [PubMed] [Google Scholar]
- Urabe M. Ding C. Kotin R.M. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum. Gene Ther. 2002;13:1935–1943. doi: 10.1089/10430340260355347. [DOI] [PubMed] [Google Scholar]
- Vincent K.A. Piraino S.T. Wadsworth S.C. Analysis of recombinant adeno-associated virus packaging and requirements for rep and cap gene products. J. Virol. 1997;71:1897–1905. doi: 10.1128/jvi.71.3.1897-1905.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.S. Khuntirat B. Qing K. Ponnazhagan S. Kube D.M. Zhou S. Dwarki V.J. Srivastava A. Characterization of wild-type adeno-associated virus type 2-like particles generated during recombinant viral vector production and strategies for their elimination. J. Virol. 1998;72:5472–5480. doi: 10.1128/jvi.72.7.5472-5480.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway A. Deru W. Prentice H.G. Anderson R. Construction of adeno-associated virus packaging plasmids and cells that directly select for AAV helper functions. J. Virol. Methods. 2003;114:1–10. doi: 10.1016/j.jviromet.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Xiao X. Li J. Samulski R.J. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z. Duan D. Engelhardt J.F. Mechanism of Recombinant Adeno-associated Virus Transduction. Hodder Arnold; London: 2006. [Google Scholar]
- Ye C. Qiu J. Pintel D.J. Efficient expression of the adeno-associated virus type 5 p41 capsid gene promoter in 293 cells does not require Rep. J. Virol. 2006;80:6559–6567. doi: 10.1128/JVI.00387-06. [DOI] [PMC free article] [PubMed] [Google Scholar]