Figure 5. BRCA1-associated differential methylation of the ZEB1 binding locus controls ZEB1 repressor binding and TAp73 expression.
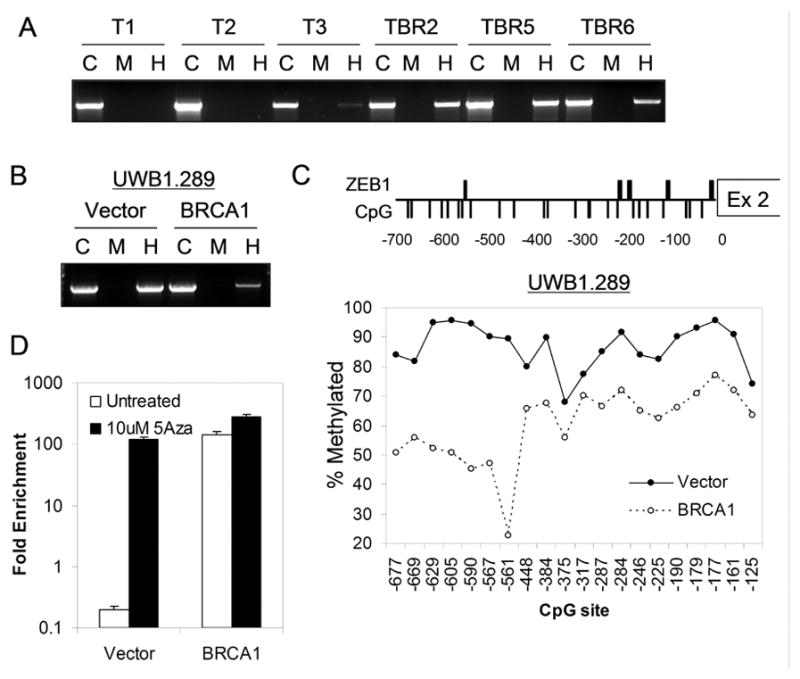
(A) Hypermethylation of the ZEB1 binding locus in murine BRCA1-deficient (TBR) versus wild-type (T) ovarian carcinoma cells. Genomic DNA was digested with MspI (M), HpaII (H), or undigested control (C) followed by PCR amplification of a 1.5kb fragment encompassing the ZEB1 binding locus. A ratio of H/C approaching 1:1 indicates hypermethylation. (B) Hypermethylation of BRCA1-deficient (Vector) versus BRCA1-reconstituted (BRCA1) human ovarian carcinoma cells. (C) Bisulfite sequencing showing consistent CpG hypermethylation across the ZEB1 binding locus in BRCA1-deficient human cells. Top, schematic showing putative ZEB binding motifs (thick bars) and CpG motifs (thin bars). Bottom, quantitation of methylation (see methods) for each CpG motif. The X-axis indicates positions of CpGs relative to exon 2. (D) Demethylation promotes ZEB1 binding in BRCA1-deficient cells. Fold-enrichment by ChIP 24 hours post 5-Aza treatment, using a ZEB1-specific versus control antibody, assayed by QPCR for fragment H5 (Figure 4D) in UWB1.289 cells.
