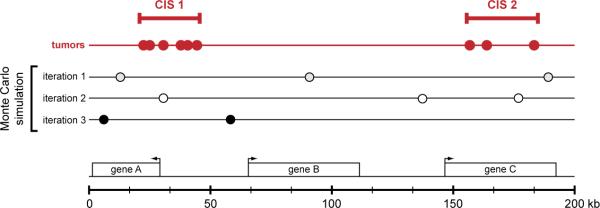
Figure 3.
Identification of common insertion sites in SB-induced tumors. High throughput PCR-based methods have recently been developed to identify a large number of transposon-induced mutations in individual SB-induced tumors. The driver mutations causally linked to transformation are identified as common insertion sites (CISs) — regions of the genome that harbor transposon insertions in multiple independent tumors. Currently, a Monte Carlo simulation is used to model random transposon insertion within the mouse genome to mimic the number of tumors and insertion events observed within each experiment. The cumulative results of thousands of interations of the Monte Carlo simulation are used to identify non-random clusters of transposon insertions in the tumor data set. These regions are then defined as CISs. The figure graphically depicts this analysis to identify CISs within a region of the genome containing three hypothetical genes (geneA-C) shown below. Each circle represents an independent transposon insertion in this 200 kilobase interval. The results of three iterations of the Monte Carlo simulation are shown, and these results are used to identify CISs in the tumor data set (shown in red).