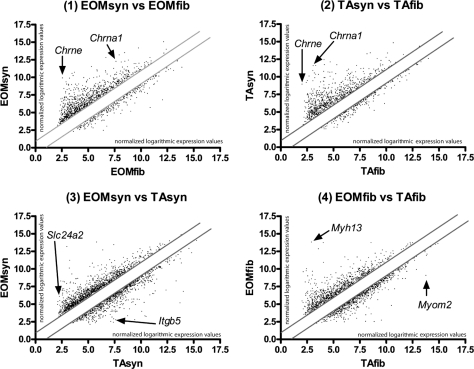
Figure 3.
Scattergram analysis of expression profiling. The four scattergraphs represent the four sets of expression profiling experiments undertaken in this study. Axes show logarithmic expression levels of each gene on a linear scale. The graphs compare the expression profiling data sets for the following independent samples/conditions: (1) EOMsyn versus EOMfib, (2) TAsyn versus TAfib, (3) EOMsyn versus TAsyn, and (4) EOMfib versus TAfib. Solid gray lines: a twofold cutoff. Genes situated farthest from the diagonal showed greatest expression differences between the two expression profiling data sets compared in each graph. (1, 2) The synaptic markers Chrna1 and Chrne upregulated in both EOMsyn and TAsyn. Examples of differentially expressed genes in (3) EOMsyn versus TAsyn are Slc24a2 and Itgb5. Myh13 was upregulated in EOMfib; Myom2 was downregulated in EOMfib (4).