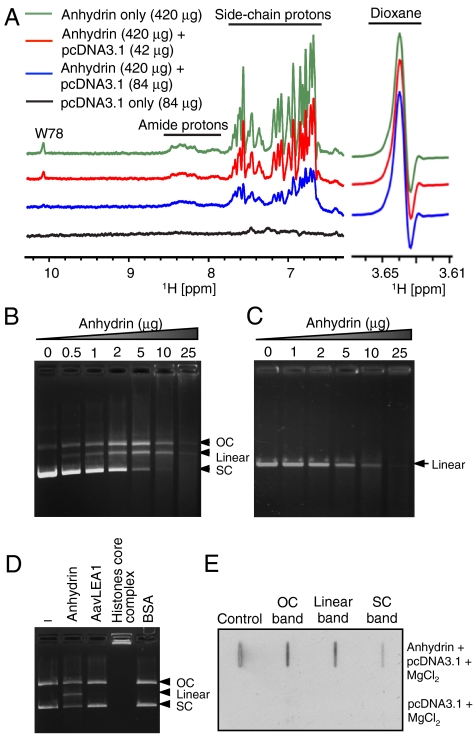
Fig. 5.
Anhydrin binds and digests DNA. (A) The 1H-NMR spectrum of free anhydrin (420 μg, green) shows signals arising from backbone amide and side-chain protons that are diminished upon titration with plasmid DNA in a progressive manner (red 42 μg, blue 84 μg DNA added). The maintenance of height and width of the resonant peak of dioxane (right, 3.64 ppm), shown as reference, indicates instrument conditions were constant throughout and that perturbation of anhydrin NMR signals was caused by interaction with DNA. (B) pcDNA3.1 (400 ng) and (C) linear DNA (mCherry gene; 200 ng) incubated with increasing concentrations of anhydrin. (D) Activity of anhydrin compared to His6-AavLEA1, core histones and BSA. (E) Slot-blot analysis of the different forms of pcDNA3.1 digested with anhydrin (top) or untreated pcDNA3.1 (bottom) after probing with Flag antibody; “Control” is purified anhydrin. 200 ng pcDNA3.1 and 2 μg anhydrin were used above, unless otherwise stated. OC, open-circular DNA; SC, supercoiled DNA.