Abstract
The Ashkenazi Jewish (AJ) population has long been viewed as a genetic isolate, yet it is still unclear how population bottlenecks, admixture, or positive selection contribute to its genetic structure. Here we analyzed a large AJ cohort and found higher linkage disequilibrium (LD) and identity-by-descent relative to Europeans, as expected for an isolate. However, paradoxically we also found higher genetic diversity, a sign of an older or more admixed population but not of a long-term isolate. Recent reports have reaffirmed that the AJ population has a common Middle Eastern origin with other Jewish Diaspora populations, but also suggest that the AJ population, compared with other Jews, has had the most European admixture. Our analysis indeed revealed higher European admixture than predicted from previous Y-chromosome analyses. Moreover, we also show that admixture directly correlates with high LD, suggesting that admixture has increased both genetic diversity and LD in the AJ population. Additionally, we applied extended haplotype tests to determine whether positive selection can account for the level of AJ-prevalent diseases. We identified genomic regions under selection that account for lactose and alcohol tolerance, and although we found evidence for positive selection at some AJ-prevalent disease loci, the higher incidence of the majority of these diseases is likely the result of genetic drift following a bottleneck. Thus, the AJ population shows evidence of past founding events; however, admixture and selection have also strongly influenced its current genetic makeup.
Keywords: genetic isolate, heterozygosity, identity-by-descent, linkage disequilibrium
The Ashkenazi Jewish (AJ) population has long been viewed as a genetic isolate, kept separate from its European neighbors by religious and cultural practices of endogamy (1). Population isolates are frequently used in genetic research, as such groups are presumed to have reduced genetic diversity, along with increased frequencies of recessive disorders, identity-by-descent (IBD), and linkage disequilibrium (LD) as the result of founder events and population bottlenecks (2, 3). Accordingly, the AJ population is often the subject of Mendelian and complex disease studies, although evidence that the AJ population carries all of the hallmarks of a genetic isolate has not been fully established.
The most compelling genetic evidence of founder events in the AJ population is the elevated frequency of at least 20 rare recessive diseases attributed to genetic drift following bottlenecks. Coalescence times ascribed to founder mutations for some of these diseases correspond well with historical migrations or episodes of extreme persecution, supporting the argument for genetic drift (1, 4, 5). Strong evidence of founder events also comes from studies of mitochondrial DNA (mtDNA), which show significantly less diversity of mtDNA haplotypes among the AJ population (6, 7). Y-chromosome studies also indicate only a low amount of admixture with neighboring Europeans (8–10). Additionally, some reports have measured higher LD in the AJ genome compared with reference populations (11, 12). Furthermore, a recent study showed increased IBD in Jewish Diaspora populations, including the AJ population, in support of a bottleneck (13).
Despite the evidence for founder events, questions still remain whether population bottlenecks and genetic isolation can account for the current genetic structure of the AJ population. For example, some have posited that selection can explain the increase in rare recessive disorders, arguing for a heterozygous advantage for the mutant alleles (14–16). Moreover, Y-chromosome studies, in contrast to mtDNA results, reveal that Y-chromosome diversity in the AJ population is comparable to their non-Jewish European neighbors (8, 17). Admixture estimates using markers at a few autosomal loci or based on STRUCTURE clustering results have also shown much higher European admixture than reflected in the Y chromosome (13, 18, 19). Furthermore, recent studies have found no increase in LD at short distances between markers and suggest increased heterozygosity compared with Europeans, concluding that the AJ population is likely an older and larger population that is distinguished by its Middle Eastern origin, rather than the effect of population bottlenecks (20–22).
To better understand the genome-wide genetic structure of the AJ population and search for genetic signatures of founder events, we genotyped 471 unrelated AJ individuals at 732k autosomal SNPs. Our analysis of this large cohort clarifies many inconsistencies described above, establishing that the AJ population has had substantial admixture with European populations, increasing its genetic diversity and LD, yet maintaining a significant level of founder haplotypes identical-by-descent. In addition, we applied extended haplotype tests for detecting regions under positive selection and, although this does not account for most AJ-prevalent disorders, we did identify several regions of putative selection in the AJ genome.
Results and Discussion
Genetic Diversity and Linkage Disequilibrium.
It is well established that populations that have been isolated for extended periods show two major patterns of diversity: (i) low genetic variation and (ii) high LD (2, 3). To compare the genetic diversity of the AJ population with their European neighbors, we merged our genotype data for 471 Ashkenazi Jews with two large European cohorts, a continental European (Euro) cohort of 1,705 individuals (242k overlapping SNPs), and a European American (EA) cohort of 1,251 individuals (732k overlapping SNPs). Consistent with recent reports (13, 20, 23–25), principal component analysis (PCA) using these combined datasets confirmed that the AJ individuals cluster distinctly from Europeans, aligning closest to Southern European populations along the first principal component, suggesting a more southern origin, and aligning with Central Europeans along the second, consistent with migration to this region (Fig. S1).
To explore the amount of genetic variation within the AJ and European populations, we first measured the mean heterozygosity. Surprisingly, we found a higher level of heterozygosity among AJ individuals compared with Europeans (P < 1e-40) (Table 1), confirming speculation made in one recent report and a trend seen in another (20, 22). Although this difference may appear small, it is highly statistically significant because of the large number of individuals and markers analyzed, even after pruning SNPs that are in high LD. The higher diversity in the AJ population was paralleled by a lower inbreeding coefficient, F, indicating the AJ population is more outbred than Europeans, not inbred, as has long been assumed (P < 1e-7) (Table 1). The greater genetic variation among the AJ population was further confirmed using a pairwise identity-by-state (IBS) permutation test, which showed that average pairs of AJ individuals have significantly less genome-wide IBS sharing than pairs of EA or Euro individuals (empirical P value < 0.05). Thus, our results show that the AJ population is more genetically diverse than Europeans.
Table 1.
Genetic diversity in Ashkenazi Jews and Europeans
| Pop | LD pruned | SNPs | HET(Exp) | HET(Obs) | F |
| AJ | No | 242k | 0.2296 | 0.2298** | −0.0011* |
| Euro | No | 242k | 0.2262 | 0.2257 | 0.0022 |
| AJ | Yes | 107k | 0.2390 | 0.2394** | −0.0014* |
| Euro | Yes | 107k | 0.2363 | 0.2358 | 0.0020 |
| AJ | No | 732k | 0.2705 | 0.2714** | −0.0033* |
| EA | No | 732k | 0.2696 | 0.2697 | −0.0004 |
| AJ | Yes | 207k | 0.2518 | 0.2527** | −0.0038* |
| EA | Yes | 207k | 0.2510 | 0.2511 | −0.0005 |
Expected heterozygosity, HET(Exp), observed heterozygosity, HET(Obs), and inbreeding coefficient, F, for the AJ, Euro, and EA populations. One SNP of a pair was pruned out of the dataset if the pair was in high LD, defined as pairwise r2 > 0.5. The shading of rows highlights the AJ population with the corresponding SNP-matched European population.
**P < 1e-40 and *P < 1e-7 for AJ compared with its paired European population (t test).
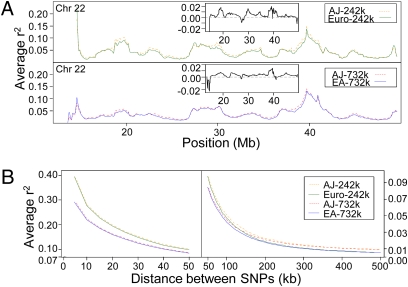
We next compared LD between populations by calculating the average r2 in sliding windows across the genome and found that LD is consistently higher in the AJ population, with only occasional regions of greater LD in Europeans (Fig. 1A). Of note, for this and all subsequent analyses, we limited the European cohorts to 471 individuals to avoid any sample size differences with the AJ population. Analysis of LD decay also demonstrates stronger LD at all distances between SNPs (Fig. 1B), contrary to a recent study that found less LD at short distances (21), illustrating the value of a larger sample size for LD calculations. The higher LD was confirmed by using a binomial test to show that significantly more SNP pairs had higher r2 in the AJ population when directly compared with the same SNP pair in the European populations (P < 9.22e-15) (Table S1). Similar results were also seen when D′ was used to measure LD.
Fig. 1.
LD in Ashkenazi Jews compared with Europeans. (A) A 1.7-Mb sliding window shifting 100 kb along chromosome 22. (Upper) AJ and Euro populations; (Lower) AJ and EA populations. For each window, the average r2 was calculated between all pairs of SNPs within 500 kb of each other. The Insets show the AJ r2 minus the Euro or EA r2, respectively. (B) The decay of LD across all of the autosomes was assessed by grouping pairs of SNPs according to the distance separating them (0–5 kb, 5–10 kb, …, 495–500 kb) and then plotting the average r2 of each bin. (Left) Distances from 0 to 50 kb; (Right) distances from 50 to 500 kb. Note the scales on the y axis are different for the Left and Right.
We also compared the genome-wide haplotype structure between the AJ and European populations using a haplotype modeling algorithm (26), which models phased haplotypes as edges that pass through nodes at each SNP across the genome. The number of nodes in the model is correlated to the genetic variation, and the number of edges per node is inversely correlated to the haplotype length. Using this model, we found that the AJ population has a greater number of nodes (0.88–1.11% more) but fewer edges per node (3.82–4.76% fewer) compared with the Europeans (P < 1e-50) (Table S2), indicating both higher genetic variation and longer haplotypes in the AJ population, consistent with our previous results.
Although the elevated LD and longer haplotypes could be the result of severe bottlenecks and founder events, such events would not account for the increased genetic diversity. However, increased LD can also arise from the admixture of genetically distinct populations (27–30), which could also explain the elevated diversity. Therefore, our data suggest that the higher diversity and LD observed in the AJ population may be the result of admixture rather than founder effects. An alternative explanation is that the AJ population simply arose from a more genetically diverse Middle Eastern founder population and, therefore, its diversity is reduced relative to the founder but not relative to the host Europeans. To address this possibility, we merged our autosomal AJ genotype data with data from the Human Genome Diversity Panel (HGDP) (31), with 168k overlapping SNPs. We removed SNPs in high LD and measured the mean heterozygosity per locus across the combined Middle Eastern populations (Bedouin, Palestinian, and Druze) and found that the AJ population had higher heterozygosity (0.3121 vs. 0.3053, P < 1e-23). Other reports showing no increased heterozygosity in the AJ relative to Middle Eastern populations (13, 22) were probably limited by lower AJ sample sizes, which our dataset overcomes. Thus, the increased genetic diversity and LD appear consistent with admixture rather than founding effects.
Admixture.
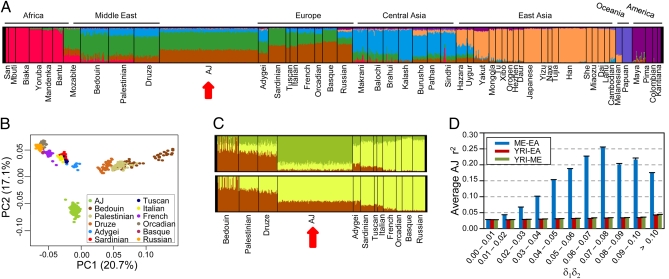
To evaluate admixture in the AJ population, we investigated the similarity between AJ and HGDP populations using PCA as well as a population clustering algorithm (32). Both analyses show that AJ individuals cluster between Middle Eastern and European populations (Fig. 2 A and B and Fig. S2A), corroborating other recent reports (13, 20, 22, 23, 25). Interestingly, our population clustering reveals that the AJ population shows an admixture pattern subtly more similar to Europeans than Middle Easterners (Fig. 2 A and C, Lower), while also verifying that the Ashkenazi Jews possess a unique genetic signature clearly distinguishing them from the other two regions (Fig. 2C, Upper). The fixation index, FST, calculated concurrently to the PCA, confirms that there is a closer relationship between the AJ and several European populations (Tuscans, Italians, and French) than between the AJ and Middle Eastern populations (Fig. S2B). This finding can be visualized with a phylogenetic tree built using the FST data (Fig. S2C), showing that the AJ population branches with the Europeans and not Middle Easterners. Two recent studies performing PCA and population clustering with high-density SNP genotyping from many Jewish Diaspora populations, both showed that of the Jewish populations, the Ashkenazi consistently cluster closest to Europeans (13, 25). Genetic distances calculated by both groups also show that the Ashkenazi are more closely related to some host Europeans than to the ancestral Levant (13, 25). Although the proximity of the AJ and Italian populations could be explained by their admixture prior to the Ashkenazi settlement in Central Europe (13), it should be noted that different demographic models may potentially yield similar principal component projections (33); thus, it is also consistent that the projection of the AJ populations is primarily the outcome of admixture with Central and Eastern European hosts that coincidentally shift them closer to Italians along principle component axes relative to Middle Easterners. Taken as a whole, our results, along with those from previous studies, support the model of a Middle Eastern origin of the AJ population followed by subsequent admixture with host Europeans or populations more similar to Europeans. Our data further imply that modern Ashkenazi Jews are perhaps even more similar with Europeans than Middle Easterners.
Fig. 2.
Admixture and its relationship with LD. (A–C) Ancestral population clustering and PCA were performed using the combined AJ and HGDP populations. The AJ population was divided into three random subgroups of 157 individuals to better match the population size of the Middle Eastern and European populations in the HGDP dataset. The data shown represent one subgroup, and all three had similar results. (A) Population clustering analysis of AJ and all HGDP individuals for seven, K = 7, theoretical ancestral populations. Each color represents a different ancestral population and each vertical line represents a single individual. The proportion of each color within an individual signifies the fraction of ancestry derived from the color's ancestral population. The red arrow highlights the AJ population. (B) PCA performed using only the AJ, Middle Eastern, and European HGDP populations. (C) Population clustering analysis with K = 3 (Upper) or K = 2 (Lower) ancestral populations, performed using only the AJ, Middle Eastern, and European HGDP populations. (D) The correlation between admixture and LD was determined by plotting the average r2 for all SNP pairs within the given intervals of δ1δ2, where δ1 is the allele frequency difference between the founding populations at locus 1, and δ2 is the frequency difference at locus 2. The legend indicates the populations representing the founding populations: Middle Eastern (ME), European American (EA), or Yoruba in Ibadan, Nigeria (YRI). Error bars show SEM.
To quantify the level of admixture within the AJ genome given the model of a Middle Eastern origin and European admixture, we applied a likelihood method (34) to differentiate the relative ancestry of each locus across the genome. We used the combined Palestinian and Druze populations to represent the Middle Eastern ancestor and tested three different European groups as the European ancestral population (SI Materials and Methods). Using these proxy ancestral populations, we calculated the amount of European admixture in the AJ population to be 35 to 55%. Previous estimates of admixture levels have varied widely depending on the chromosome or specific locus being considered (18), with studies of Y-chromosome haplogroups estimating from 5 to 23% European admixture (8, 9). Our higher estimate is in part a result of the use of different proxies for the Jewish ancestral population. Our analysis used the Middle Eastern population frequencies as the putative Jewish ancestor, similar to a previous approach (18), whereas the studies of Y-chromosome admixture used a combination of several Jewish Diaspora populations. Our calculations will have overestimated the level of admixture if the true Jewish ancestor is genetically closer to Europeans than Middle Easterners; however, using the Jewish Diaspora populations as the reference Jewish ancestor will naturally underestimate the true level of admixture, as the modern Jewish Diaspora has also undergone admixture since their dispersion. Furthermore, because our analysis incorporates data from considerably more markers across all of the autosomes, thus including both male and female contributions to admixture, we believe our estimate is closer to the true level of admixture. Recent STRUCTURE analysis of the entire Jewish Diaspora estimated that the Ashkenazi and Syrian Jewish populations have between 20 and 40% European admixture (13). Our estimate overlaps this assessment, although we avoided a similar approach because STRUCTURE-like algorithms model hypothetical ancestral populations that likely never existed in reality (35).
Next, we wanted to consider if admixture could account for the increased LD in the AJ population. Admixture between genetically differentiated populations gives rise to an increase in LD proportional to δ1δ2, where δ1 is the allele frequency difference between the founding populations at locus 1, and δ2 is the frequency difference at locus 2 (27, 29, 30). Admixture LD decays within a few generation at long distances (>20 cM) but decays slowly at short distances (< 10 cM) (27, 29), allowing us to detect admixture LD in the AJ genome even if admixture occurred early in its history. We, therefore, tested if allele frequency differences between the Middle Eastern and European populations correlated with elevated AJ LD. Indeed, we see that the increase in δ1δ2 coincides dramatically with increased LD in the AJ population (Fig. 2D). Using an alternative ancestral population, the Yoruba (YRI), which has even greater allele frequency differences, does not show a similar trend, confirming that admixture between Middle Eastern and European populations contributes to the high LD seen in the AJ population. Models proposed by Kruglyak (36) imply that a population isolate would have to undergo an extreme bottleneck to see a significant increase in LD. Admixture models, on the other hand, reveal that LD can be elevated even when admixture rates are low (27–29). Therefore, our data suggest that the elevated LD in the AJ population has arisen primarily as the result of admixture rather than founder effects.
Identity-by-Descent.
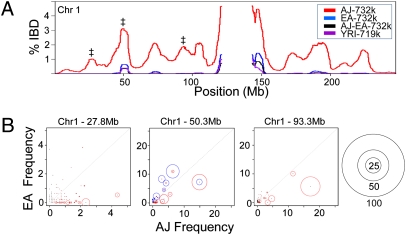
Another genetic hallmark of population isolates is elevated IBD. We searched for regions of IBD using a hashing and extension algorithm (37) and discovered significantly more segments of IBD in the AJ population compared with Europeans or an out-group population, YRI (P < 1e-10) (Table S3). Plotting the frequency of IBD across the genome reveals that most loci are usually shared by less than 5% of AJ pairs (Fig. 3A and Fig. S3A). The exceptions to this are the pericentromeric regions that often show a high frequency of sharing in all populations, possibly because of low SNP density or other factors. A recent study by Atzmon et al. (13) also reported higher IBD in a small AJ cohort; however, we found considerably more IBD in our AJ population (4.5 segments per pair vs. 1.6 segments per pair), even when the IBD segments were similarly filtered (Table S3, Bottom). Given our much larger AJ cohort, as well as the availability of trios to help in haplotype phasing, our results are likely more precise. When we plotted the IBD length decay and maximum IBD length distribution we did see similar trends to Atzmon et al., consistent with a bottleneck in the AJ history (Fig. S3 B and C). Furthermore, the increased IBD in the AJ population was uniform across the genome, indicating it was not the byproduct of a few loci under selection, but likely the outcome of founder events and the persistence of long founding haplotypes.
Fig. 3.
IBD and frequency of shared haplotypes. (A) The percentage of IBD for all pairs of individuals within the AJ, EA, and YRI populations is plotted for chromosome 1. Pairs consisting of one Ashkenazi Jew with one European American (AJ-EA) were also analyzed. The symbol ‡ mark the three loci whose haplotype frequencies are analyzed in B. (B) The 100-SNP haplotype blocks centered at three loci marked in A were analyzed for their frequency in the AJ and EA population. Red circles represent the haplotypes identified with IBD in the AJ population and the size of the circle indicates the frequency that the haplotype is shared (i.e., 50 indicates that the given haplotype is shared by half of the AJ pairs with IBD at that locus). The scale of the circles is given on the far right. (Center) Blue circles represent the haplotypes with IBD in the EA population.
Because less than 5% of the AJ pairs share IBD at any given locus, this implies that the long founding haplotypes are present at low frequency in the AJ population. To explore this, we measured the relative IBD-sharing frequencies of long haplotypes at three loci on chromosome 1 (Fig. 3B). This analysis confirms that many long haplotypes sharing IBD are found at low frequency in the AJ population and are not present in the EA population, presumably representing the existence of older founder haplotypes. The presence of multiple haplotypes contributing to the overall level of IBD at a given locus also confirms that the IBD is not the result of selection of a single haplotype. Additionally, some haplotypes sharing IBD in the AJ population are also present in lower frequency in the EA population, which may be evidence of early admixture. Taken together, our IBD results support the existence of founder effects in the history of the AJ population, as well as admixture with European neighbors.
Positive Selection.
Positive selection at disease loci is an alternative explanation to genetic drift that could account for the prevalence of AJ diseases (14–16). Our study is unique in applying extended haplotype tests, the integrated haplotype score (iHS) and cross-population extended haplotype homozygosity test (XP-EHH) (38, 39), to the AJ population. Both tests are designed to uncover selected alleles with higher frequency than expected relative to their haplotype length. The iHS method has greater power to detect selected alleles at intermediate frequency, and the XP-EHH test has greater power when selected alleles approach fixation in one population relative to another (38). Importantly, the XP-EHH test normalizes for differences in genome-wide haplotype length to account for demographic differences between populations, allowing us to directly compare signals of selection in the AJ and European populations (38). Our IBD analysis suggests that the AJ population shares long haplotypes as the result of founder events or bottlenecks. However, because of the low frequency of these long shared haplotypes, they should not interfere with identifying true regions under strong selective pressure. Furthermore, these long-range haplotype tests are designed to test the hypothesis of neutrality, so rejection suggests that selection has likely occurred, but failure to reject does not mean that selection is absent, implying that undetected loci under selection exist.
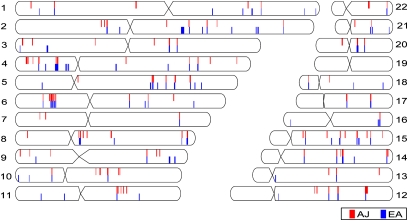
We first performed the iHS test separately for the AJ and EA populations and we observed similar magnitudes of integrated haplotype scores between the populations, indicating that the frequency of founding haplotypes in the AJ population did not substantially alter the extended haplotype scores. We calculated the fraction of SNPs with standardized |iHS| > 2 for nonoverlapping, 40-SNP windows across the genome, an approach shown to be more powerful for detecting true regions of selection rather than relying on iHS scores for single SNPs (39). Comparing the top iHS windows in both populations reveals many regions of putative selection in common, including several regions previously implicated as being under selective pressure (Fig. 4 and Table S4). Approximately half of the top 1% of iHS windows in each population are shared by both. This finding is consistent with a recent report that found large overlap between selected regions in Middle Eastern and European populations (40).
Fig. 4.
The top 1% of iHS-selected regions. Each chromosomal ideogram shows the position of the top 1% of iHS windows in the AJ population, red, and the EA population, blue.
To explore whether regions of selection in the AJ population included any loci of known Ashkenazi diseases, we examined 21 disease- and cancer-susceptibility loci with known mutations found at higher frequency in the Ashkenazi population. Only 6 of the 21 genes fell in or near (within 500 kb) the top 5% of the AJ iHS windows (Table 2). Among these is the Tay-Sachs disease gene, HEXA, whose selection has been widely debated (4, 5, 14–16) and was found ~400 kb downstream of a window on chromosome 15 identified in the top 1% of the AJ iHS hits. Although none of the SNPs interrogated immediately adjacent to the HEXA locus showed elevated iHS signals, it is possible that the nearby region may contain regulatory elements under selection that affect HEXA expression. Cochran et al. (14) speculated that selection of many of the AJ-prevalent disease loci, especially the lysosomal diseases, conferred an increase in intelligence that was necessary historically for the AJ economic survival. Our data shows evidence of strong selection at or near only six disease loci, including only one out of the four AJ-prevalent lysosomal storage diseases, thus arguing that most AJ disease loci are not under strong positive selection, but rather rose to their current frequency through genetic drift after a bottleneck. However, we cannot exclude the possibility that selection of some AJ disease loci are outside the limits of detection by the extended haplotype tests, which are known to have less power to detect selection of lower frequency alleles (38, 41).
Table 2.
Overlap of AJ disease loci and regions of selection
| Disease | Carrier freq. | Gene | Selected region | |
| AJ-prevalent disease | ||||
| Bloom syndrome | 1/100 | BLM | No | |
| Canavan | 1/41 | ASPA | No | |
| Congenital adrenal hyperplasia | 1/10 | CYP21A2 | Yes | |
| Factor XI deficiency | 1/19 | F11 | No | |
| Familial dysautonomia | 1/30 | IKBKAP | No | |
| Familial nonsyndromic deafness | 1/25 | GJB2 | No | |
| Fanconi anemia C | 1/90 | FANCC | Yes | |
| Gaucher Type 1 | 1/18 | GBA | No | |
| Glycogen storage disease type 1a | 3/200 | G6PC | No | |
| HMPS1 (colorectal cancer) | ? | CRAC1 | No | |
| Mucolipidosis Type IV | 1/50 | MCOLN1 | No | |
| Niemann-Pick Type A | 1/80 | SMPD1 | No | |
| Tay-Sachs | 1/30 | HEXA | Yes | |
| Torsion dystonia | 1/2,000 | DYT1 | No | |
| AJ-Prevalent alleles | ||||
| Breast/Ovarian cancer | 1/100 | BRCA1 | No | |
| Breast/Ovarian cancer | 1/100 | BRCA2 | Yes | |
| Colorectal cancer | 1/17 | APC | Yes | |
| Familial hypercholesterolemia | 1/56 | LDLR | No | |
| Familial hyperinsulinism | 1/89 | ABCC8 | Yes | |
| HNPCC1 (colorectal cancer) | 1/100 | MSH2 | No | |
| Maple syrup urine disease | 1/113 | BCKDHB | No | |
Table compiled from Risch et al. (4), Kedar-Barnes and Rozen (56), Ostrer, H (1), and Charrow, J (57), showing disease and cancer susceptibility loci at increased frequency in the Ashkenazi Jews. The bottom portion of the table lists diseases for which specific mutant alleles are at higher frequency in the AJ population, although the overall disease incidence is similar in other populations. Being in a selected region was defined as falling in or near (within 500 kb of) the top 5% of iHS windows.
To uncover other loci that show evidence of stronger selection in either the AJ or EA populations, we identified regions found in the top 1% of iHS hits in one population but not the other (Fig. 4 and Table S5), as well as directly comparing populations using the XP-EHH test (Table 3). Both tests indicated that the strongest signal of selective pressure unique to Europeans was at the lactase locus, LCT, which we found to be under strong selective pressure in the EA population, but showed no signs of selection in our data for the AJ population. Multiple studies have found that the “lactase-persistence” allele at the LCT locus was selected for in Northern Europeans, with the selective sweep presumably occurring at the time of the domestication of cattle 2,000 to 20,000 y ago (42, 43). The absence of this allele in our data would suggest that the selective sweep was complete before the Ashkenazi establishment in Europe. Moreover, the prevalence of lactase deficiency in Ashkenazi Jews has been estimated at 60 to 80% (44), further corroborating the lack of selection for the LCT locus in the AJ population.
Table 3.
Top XP-EHH regions of selection
| Chr: region (Mb hg18) | Max |XP-EHH| | Pop selected | Genes (number) |
| 1: 160.36–160.54 | 4.53 | EA | NOSIAP (1) |
| 2: 134.63–137.29 | 13.36 | EA | CCNT2, RAB3GAP1, LCT (13) |
| 2: 237.19–237.34 | 4.62 | EA | None (0) |
| 3: 10.59–10.86 | 5.07 | EA | SLC6A11, LOC285370 (2) |
| 3: 11.59–11.71 | 5.07 | EA | VGLL4 (1) |
| 4: 30.66–31.01 | 4.86 | EA | PCDH7 (1) |
| 5: 56.95–57.29 | 4.65 | AJ | None (0) |
| 5: 112.64–113.18 | 4.73 | AJ | MCC, YTHDC2 (2) |
| 6: 31.35–32.47 | 5.95 | AJ | TNF, CYP21A2, SLC44A4 (60) |
| 6: 156.46–156.76 | 4.77 | AJ | None (0) |
| 8: 49.2–49.31 | 5.04 | EA | None (0) |
| 8: 52.12–52.36 | 4.74 | EA | None (0) |
| 8: 55.54–55.97 | 4.55 | AJ | RP1 (1) |
| 8: 80.25–80.73 | 4.69 | EA | STMN2 (1) |
| 11: 12.84–12.93 | 4.78 | AJ | TEAD1 (1) |
| 11: 79.65–79.79 | 5.05 | AJ | None (0) |
| 12: 109.84–111.85 | 5.95 | AJ | ATXN2, ALDH2, TRAFD1 (18) |
| 13: 98.06–98.21 | 4.54 | EA | SLC15A1 (1) |
| 15: 46.73–47.17 | 5.18 | AJ | SHC4, EID1, SECISBP2L (4) |
| 18: 73.91–74.11 | 4.48 | AJ | None (0) |
The XP-EHH test directly compared the AJ and EA populations for regions that show stronger selection in one population relative to the other. The top 10 selected regions for each population are reported. A subset of genes in each interval is listed, with the total number in that region in parentheses.
The strongest signal of selection unique to the AJ population was on chromosome 12 (110.6–111.72 Mb hg18). This locus was not found to be under selection in either the EA population or among reported selected regions of the HGDP Middle Eastern populations (40), making it an apparent Jewish-specific selected locus. Interestingly, this locus is also within a region that was recently identified as having high IBD across many Jewish Diaspora populations (13). This region contains 18 genes, including the mitochondrial aldehyde dehydrogenase gene, ALDH2, which is part of the major oxidative pathway of alcohol metabolism. Genetic variation within ALDH2 has been shown repeatedly to affect alcohol dependence (45). Intriguingly, the AJ population has long been known to have lower levels of alcoholism than other groups (16, 46), with one study showing that Jewish males have a 2.5-fold lower lifetime rate of alcohol abuse/dependence compared with non-Jews (47). In his analysis of the historical record, Keller (46) concluded that drunkenness was common among Jews before their Babylonian captivity but that drunkenness vanished around the time they returned to Israel in 537 B.C.E. This dramatic shift and persistence of low alcoholism rates in modern Jews has largely been attributed to social and religious practices (16, 46), with little support for a biological explanation. Our results, together with a recent study showing that variation in the ALDH2 promoter affects alcohol absorption in Jews (48), now suggest that genetic factors and selective pressure at the ALDH2 locus may have contributed to the low levels of alcoholism. The mechanism driving selection of the ALDH2 locus is unknown, but a plausible target of selection also within this selected region is the TRAFD1/FLN29 gene, which is a negative regulator of the innate immune system, important for controlling the response to bacterial and viral infection (49). TRAFD1/FLN29 may have conferred a selective advantage in the immune response to a pathogen, perhaps near the time that the Jews returned to Israel from their Babylonian captivity. Despite the unclear selective mechanism, this remains a remarkable example of a putatively selected region accounting for a known population phenotype.
Materials and Methods
More detailed methods are described in SI Materials and Methods and Dataset S1.
Genotype Data.
Unrelated Ashkenazi Jews were genotyped using the Affymetrix 6.0 array, with 471 individuals and 732K SNPs passing quality control filters. Genotype data for all other populations were obtained from previously published studies or publicly available sources.
Principal Component Analysis, FST, and Phylogenetic Tree Building.
PCA was performed using smartpca in the EIGENSOFT software package (v 3.0) (50, 51). An FST matrix was calculated using smartpca concurrently with the PCA analysis. The unrooted phylogenetic tree built using the FST matrix was created using the FITCH program in the PHYLIP package (v 3.69) (52).
Ancestral Clustering and Locus-Specific Admixture.
The frappe algorithm (v 1.0) (32) was used to determine the ancestral population clustering. The LAMPANC algorithm was used from LAMP (v2.3) (34) to calculate the locus-specific admixture given two ancestral populations.
Genetic Diversity.
Heterozygosity (HET) and inbreeding coefficients (F) were calculated using all nonmissing genotype calls. The pairwise IBS test was run in PLINK (53).
Linkage Disequilibrium.
The r2 and D′ were calculated for all pairs of SNPs within 500 kb of each other in Haploview (v 4.1) (54).
Haplotype Phasing and Frequency Modeling.
BEAGLE (v 3.04) was used to phase haplotypes (55) and build a graphical model of haplotype frequency (26).
Identity-by-Descent.
Phased haplotypes from BEAGLE were analyzed with the GERMLINE program (v 1.4.0) (37) to identify segments of IBD (> 5 Mb, 150 SNPs).
Positive Selection.
The iHS and XP-EHH were implemented according to previous methods (38–40).
Supplementary Material
Acknowledgments
We thank M. Kayser, Erasmus University Medical Center, for use of European genotype data, and H. von Eller-Eberstein, Christian Albrechts University, for providing genotype data from the German Kiel population. The population reference sample (POPRES) and European American datasets were obtained from the database of Genotypes and Phenotypes (dbGaP). Genotyping of the POPRES sample was funded by GlaxoSmithKline and submitted by M. Nelson. Genotyping of the European-American samples was provided through the Genetic Association Information Network (GAIN) and were submitted to dbGaP by P. Gejman and the National Institute of Mental Health-funded Molecular Genetics of Schizophrenia Collaboration. We also thank the Human Genome Diversity Project for making their genotype data available. We thank all the authors of computer algorithms used in our analysis, and V. Patel for assistance in configuring them for our use. In addition, we thank members of the Warren laboratory and C. Strauss for help in reviewing the manuscript, as well as D. Cutler for helpful advice during the project. This work was supported, in part, by National Institutes of Health Grants MH080129 and MH083722 (to S.T.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004381107/-/DCSupplemental.
Data deposition: The AJ genotype data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE23636).
References
- 1.Ostrer H. A genetic profile of contemporary Jewish populations. Nat Rev Genet. 2001;2:891–898. doi: 10.1038/35098506. [DOI] [PubMed] [Google Scholar]
- 2.Arcos-Burgos M, Muenke M. Genetics of population isolates. Clin Genet. 2002;61:233–247. doi: 10.1034/j.1399-0004.2002.610401.x. [DOI] [PubMed] [Google Scholar]
- 3.Peltonen L, Palotie A, Lange K. Use of population isolates for mapping complex traits. Nat Rev Genet. 2000;1:182–190. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- 4.Risch N, Tang H, Katzenstein H, Ekstein J. Geographic distribution of disease mutations in the Ashkenazi Jewish population supports genetic drift over selection. Am J Hum Genet. 2003;72:812–822. doi: 10.1086/373882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slatkin M. A population-genetic test of founder effects and implications for Ashkenazi Jewish diseases. Am J Hum Genet. 2004;75:282–293. doi: 10.1086/423146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behar DM, et al. MtDNA evidence for a genetic bottleneck in the early history of the Ashkenazi Jewish population. Eur J Hum Genet. 2004;12:355–364. doi: 10.1038/sj.ejhg.5201156. [DOI] [PubMed] [Google Scholar]
- 7.Behar DM, et al. The matrilineal ancestry of Ashkenazi Jewry: Portrait of a recent founder event. Am J Hum Genet. 2006;78:487–497. doi: 10.1086/500307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behar DM, et al. Contrasting patterns of Y chromosome variation in Ashkenazi Jewish and host non-Jewish European populations. Hum Genet. 2004;114:354–365. doi: 10.1007/s00439-003-1073-7. [DOI] [PubMed] [Google Scholar]
- 9.Hammer MF, et al. Jewish and Middle Eastern non-Jewish populations share a common pool of Y-chromosome biallelic haplotypes. Proc Natl Acad Sci USA. 2000;97:6769–6774. doi: 10.1073/pnas.100115997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nebel A, et al. High-resolution Y chromosome haplotypes of Israeli and Palestinian Arabs reveal geographic substructure and substantial overlap with haplotypes of Jews. Hum Genet. 2000;107:630–641. doi: 10.1007/s004390000426. [DOI] [PubMed] [Google Scholar]
- 11.Service S, et al. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat Genet. 2006;38:556–560. doi: 10.1038/ng1770. [DOI] [PubMed] [Google Scholar]
- 12.Shifman S, Kuypers J, Kokoris M, Yakir B, Darvasi A. Linkage disequilibrium patterns of the human genome across populations. Hum Mol Genet. 2003;12:771–776. doi: 10.1093/hmg/ddg088. [DOI] [PubMed] [Google Scholar]
- 13.Atzmon G, et al. Abraham's children in the genome era: Major Jewish Diaspora populations comprise distinct genetic clusters with shared Middle Eastern Ancestry. Am J Hum Genet. 2010;86:850–859. doi: 10.1016/j.ajhg.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cochran G, Hardy J, Harpending H. Natural history of Ashkenazi intelligence. J Biosoc Sci. 2006;38:659–693. doi: 10.1017/S0021932005027069. [DOI] [PubMed] [Google Scholar]
- 15.Diamond JM. Human genetics. Jewish lysosomes. Nature. 1994;368:291–292. doi: 10.1038/368291a0. [DOI] [PubMed] [Google Scholar]
- 16.Goodman RM, Motulsky AG, editors. Genetic Diseases Among Ashkenazi Jews. New York: Raven Press; 1979. [Google Scholar]
- 17.Thomas MG, et al. Founding mothers of Jewish communities: Geographically separated Jewish groups were independently founded by very few female ancestors. Am J Hum Genet. 2002;70:1411–1420. doi: 10.1086/340609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavalli-Sforza LL, Carmelli D. The Ashkenazi gene pool: Interpretations. In: Goodman R, Motulsky A, editors. Genetic Diseases Among Ashkenazi Jews. New York: Raven Press; 1979. pp. 93–101. [Google Scholar]
- 19.Morton N, et al. Bioassay of kinship in populations of Middle Eastern origin and controls. Curr Anthropol. 1982;23:157–167. [Google Scholar]
- 20.Need AC, Kasperaviciute D, Cirulli ET, Goldstein DB. A genome-wide genetic signature of Jewish ancestry perfectly separates individuals with and without full Jewish ancestry in a large random sample of European Americans. Genome Biol. 2009;10:R7. doi: 10.1186/gb-2009-10-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olshen AB, et al. Analysis of genetic variation in Ashkenazi Jews by high density SNP genotyping. BMC Genet. 2008;9:14. doi: 10.1186/1471-2156-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopelman NM, et al. Genomic microsatellites identify shared Jewish ancestry intermediate between Middle Eastern and European populations. BMC Genet. 2009;10:80. doi: 10.1186/1471-2156-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian C, et al. European population genetic substructure: Further definition of ancestry informative markers for distinguishing among diverse European ethnic groups. Mol Med. 2009;15:371–383. doi: 10.2119/molmed.2009.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian C, et al. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet. 2008;4:e4. doi: 10.1371/journal.pgen.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behar DM, et al. The genome-wide structure of the Jewish people. Nature. 2010;466:238–242. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- 26.Browning SR. Multilocus association mapping using variable-length Markov chains. Am J Hum Genet. 2006;78:903–913. doi: 10.1086/503876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakraborty R, Weiss KM. Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc Natl Acad Sci USA. 1988;85:9119–9123. doi: 10.1073/pnas.85.23.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchard JK, Przeworski M. Linkage disequilibrium in humans: Models and data. Am J Hum Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephens JC, Briscoe D, O'Brien SJ. Mapping by admixture linkage disequilibrium in human populations: Limits and guidelines. Am J Hum Genet. 1994;55:809–824. [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson JF, Goldstein DB. Consistent long-range linkage disequilibrium generated by admixture in a Bantu-Semitic hybrid population. Am J Hum Genet. 2000;67:926–935. doi: 10.1086/303083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 32.Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: Analytical and study design considerations. Genet Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 33.McVean G. A genealogical interpretation of principal components analysis. PLoS Genet. 2009;5:e1000686. doi: 10.1371/journal.pgen.1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pasaniuc B, Sankararaman S, Kimmel G, Halperin E. Inference of locus-specific ancestry in closely related populations. Bioinformatics. 2009;25:i213–i221. doi: 10.1093/bioinformatics/btp197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiss KM, Long JC. Non-Darwinian estimation: My ancestors, my genes’ ancestors. Genome Res. 2009;19:703–710. doi: 10.1101/gr.076539.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet. 1999;22:139–144. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- 37.Gusev A, et al. Whole population, genome-wide mapping of hidden relatedness. Genome Res. 2009;19:318–326. doi: 10.1101/gr.081398.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabeti PC, et al. International HapMap Consortium Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pickrell JK, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabeti PC, et al. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 42.Bersaglieri T, et al. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelley JL, Swanson WJ. Positive selection in the human genome: From genome scans to biological significance. Annu Rev Genomics Hum Genet. 2008;9:143–160. doi: 10.1146/annurev.genom.9.081307.164411. [DOI] [PubMed] [Google Scholar]
- 44.Gilat T. Lactase deficiency: The world pattern today. Isr J Med Sci. 1979;15:369–373. [PubMed] [Google Scholar]
- 45.Edenberg HJ. The genetics of alcohol metabolism: Role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- 46.Keller M. The great Jewish drink mystery. Br J Addict Alcohol Other Drugs. 1970;64:287–296. doi: 10.1111/j.1360-0443.1970.tb03688.x. [DOI] [PubMed] [Google Scholar]
- 47.Levav I, Kohn R, Golding JM, Weissman MM. Vulnerability of Jews to affective disorders. Am J Psychiatry. 1997;154:941–947. doi: 10.1176/ajp.154.7.941. [DOI] [PubMed] [Google Scholar]
- 48.Fischer M, Wetherill LF, Carr LG, You M, Crabb DW. Association of the aldehyde dehydrogenase 2 promoter polymorphism with alcohol consumption and reactions in an American Jewish population. Alcohol Clin Exp Res. 2007;31:1654–1659. doi: 10.1111/j.1530-0277.2007.00471.x. [DOI] [PubMed] [Google Scholar]
- 49.Sanada T, et al. FLN29 deficiency reveals its negative regulatory role in the Toll-like receptor (TLR) and retinoic acid-inducible gene I (RIG-I)-like helicase signaling pathway. J Biol Chem. 2008;283:33858–33864. doi: 10.1074/jbc.M806923200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 52.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.69 Distributed by the author. Department of Genome Sciences, University of Washington, Seattle, WA. 2009 [Google Scholar]
- 53.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 55.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kedar-Barnes I, Rozen P. The Jewish people: Their ethnic history, genetic disorders and specific cancer susceptibility. Fam Cancer. 2004;3:193–199. doi: 10.1007/s10689-004-9544-0. [DOI] [PubMed] [Google Scholar]
- 57.Charrow J. Ashkenazi Jewish genetic disorders. Fam Cancer. 2004;3:201–206. doi: 10.1007/s10689-004-9545-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.