Abstract
Purpose of the review
To discuss potential clinical utility of the DNA sequence variants (DSVs) present in the human genome.
Recent findings
Advances in the sequencing technology have led to discovery of a very large number of DSVs in the human genome. Accordingly each genome has approximately 4 million DSVs, of which single nucleotide polymorphisms (SNPs) dominate in number (about 3 million) but the structural variations (SVs), including the copy number variants (CNVs), encompass a much larger number of the nucleotides. The biological and clinical impacts of DSVs are innate to their effect sizes and follow a gradient from negligible to drastic. DSVs responsible for single gene disorders impart the largest effect sizes, while those with small or moderate effect sizes modify phenotypic expression of the single gene disorders. In contrast, the common complex disorders result from intricate interactions of a very large number of DSVs, each imparting a modest and often clinically indiscernible effect size, with each other and with the environment factors. DSVs with large effect sizes, under certain circumstances, might have clinical utility in individualization of therapy, early diagnosis and the risk stratification. In contrast, DSVs with small effect sizes are unlikely to provide useful clinical information.
Summary
DSVs, under certain circumstances, could provide valuable information for genetic-based diagnosis, risk stratification and treatment. However, the primary utility of DSVs is in providing insight into the molecular mechanisms that govern the pathogenesis of the human diseases and applying the mechanistic insight to the cure of such disorders.
Keywords: Genetics, Genome, Next Generation DNA sequencing, Singe nucleotide polymorphisms, Structural variations, Copy number variants
Introduction
About 20 years ago when I was a post-doctoral fellow in Dr. Robert Roberts' molecular genetics laboratory and eager to learn the techniques, the advice from my mentor was: “Learn the principles of the molecular genetic techniques but don't build your academic career on techniques alone. Techniques changes but the fundamental principles stay.” Looking back to the past 3 decades of gene mapping and DNA sequencing technologies the veracity of this statement cannot be appreciated enough. The conventional technical approaches to gene mapping and DNA sequencing have all but been replaced by the newer approaches, while the fundamentals have stood the test of time. The ingenious method of DNA sequencing by synthesis for which Dr. Frederick Sanger and Dr. Walter Gilbert received the Nobel Prize in Chemistry in 1980 has remained fundamentally sound [1]. However, the approach of using radiolabeled nucleotides to label the newly synthesized DNA, dideoxynucleotides to terminate the DNA chains, polyacrylamide gel electrophoresis (PAGE) to separate the strands and autoradiography to detect the signals soon were replaced with newer methods. Fluorescent dye labeled nucleotides replaced the 32P labeled nucleotides, capillary electrophoresis replaced the slab PAGE and laser beams were used to detect the signal instead of autoradiography. The gradual increasing of the number of capillaries from a single capillary to 4-, 16-, 48- and finally 96- capillaries suddenly brought in the brave new world of genome sequencing. Soon the annotated sequence of the pooled human genomes and subsequently the first annotated sequence of a diploid genome were successfully completed by the Sanger DNA sequencing method [2, 3]. The triumphs, however, were restricted to the large-scale operations and were not – at the whole genome levels – practical in small research or clinical laboratories.
Scientific discoveries are typically incremental and only the sizes of the increments vary. A big increment in DNA sequencing technology is the development of the massively parallel DNA sequencing technique, which is categorized as a “disruptive” technology as it completely overshadows the preceding technologies. The massively parallel sequencing or deep sequencing enables sequencing of a very large number of clonal DNA strands simultaneously using the Next Generation DNA Sequencers (NexGen). The technology affords the opportunity to sequence the entire human genome in weeks and the targeted genomic regions of interest in days. The output is several Giga base pair of DNA, as it requires multiple reading of each nucleotide (20× to 100×) in order to reduce the error rates introduced by the enzymes during polymerase chain amplification (PCR) of DNA fragments and during DNA synthesis or ligation cycles. Moreover, single DNA molecule sequencing is also emerging and has the potential, by providing robust accuracy, to replace the NexGen DNA sequencers. These “disruptive” technologies have shifted the bottleneck in DNA sequencing from the high throughout capacity of the laboratories to the bioinformatics analysis of the huge amount of the data that is generated by the NexGen Sequencers. Unlike the old days, the genetic laboratory scientists no longer discover the DNA sequence variants (DSVs), Bioinformaticians do!!!
Text of Reviews
We are a very fortunate generation that has witnessed the evolution of the DNA sequencing technology from the description of the Sanger technique in 1977 [1] to the capability of sequencing the human genome for less than $5,000.00 [4]. The NexGen sequencing in conjunction with DNA target enrichment technique also afford the opportunity to sequence the entire exome in the human genome [5]. Nevertheless, as any other advancements the success in deep DNA sequencing has created new challenges that need to be resolved before the wealth of the genetic information that is offered by the NexGen DNA sequencing technology could be harnessed for the clinical use. The sequencing process is not exempt from errors. The process requires amplifications of millions of copies of fragmented DNA in parallel by the PCR. This is then followed by the sequencing reaction either through sequencing-by-synthesis or through cycles of hybridization and ligation. The first set of challenges is the technical challenges inherent to the imperfectness of the DNA polymerases and ligases used in the sequencing reactions. The relatively low error rate of the DNA amplification and sequencing enzymes is counter-balanced by the massive output of several Giga base pairs of DNA. Likewise, the impreciseness of simultaneous sequencing of clonal DNA fragments and the detection methods add to the errors. To reduce the errors each nucleotide is sequenced several times. Hence, the depth of the coverage significantly reduces the number of base call error reads. Nonetheless, even at a very low overall error rare of 10-4 per nucleotide, one would expect a very large number of erroneous nucleotides and alleles calls. Thus, bioinformatics analysis is essential for the distinction between the background noise from the real nucleotide changes and the correct calling of the bases and the alleles. Bioinformatics of deep DNA sequencing is the early stages of development and is an evolving field in need of further improvement. Robustness of bioinformatics analysis is the key element in reducing in the number of miscalls and any reduction in the number of miscalls will have drastic impact on subsequent validation sequencing, typically by the Sanger method. The above challenges are not insurmountable and likely to be resolved with further advances in the DNA sequencing technology, particularly the advancement of single DNA molecule sequencing technologies, which could potentially replace the existing NexGen DNA sequencers.
The more difficult challenge, however, is the complexity of the human genome, which has been subjected to Darwin's evolutionary pressures and geopolitical forces over billions of years and billions of meiosis. The complexity of the genome was clearly demonstrated by the published genome sequence data of Dr. Craig Venter, Dr. James Watson, an African man, a Korean man, a Han Chinese man and several exomes [3, 6-9]. Accordingly, the genome of each individual has approximately 4 million DSVs that are comprised of more than 3 million single nucleotide variants (SNPs), including about 10,000 non-synonymous, i.e., amino acid changing variants, which could potentially impart biologically functions. In addition, each genome contains several thousands structural variations (SVs), including about 200,000 small insertion/deletions (indels) and several hundreds rearrangements and duplications. SVs are fewer in the number than the SNPs. However, they encompass 75% of the variant nucleotides in the genome [3]. Some SVs involve several million nucleotides and a large number of genes [3, 10, 11]. Many SVs affect the copy number of the genes, and hence, are referred to as copy number variants (CNVs). Moreover, many DSVs in each individual's genome, whether SNPs or SVs, are often unique or rare. The data from the 5 individual genomes that have been sequenced show that approximately a quarter of the SNPs and the majority of SVs in each genome were novel. Thus, each individual genome is by and large “private” and a “personal genome”. The abundance of DNA nucleotide variants in the genome along with the personal nature of each genome pose significant challenges in discerning the variants that contribute to or cause the disease from those variants that are totally innocuous, if indeed such variants exist.
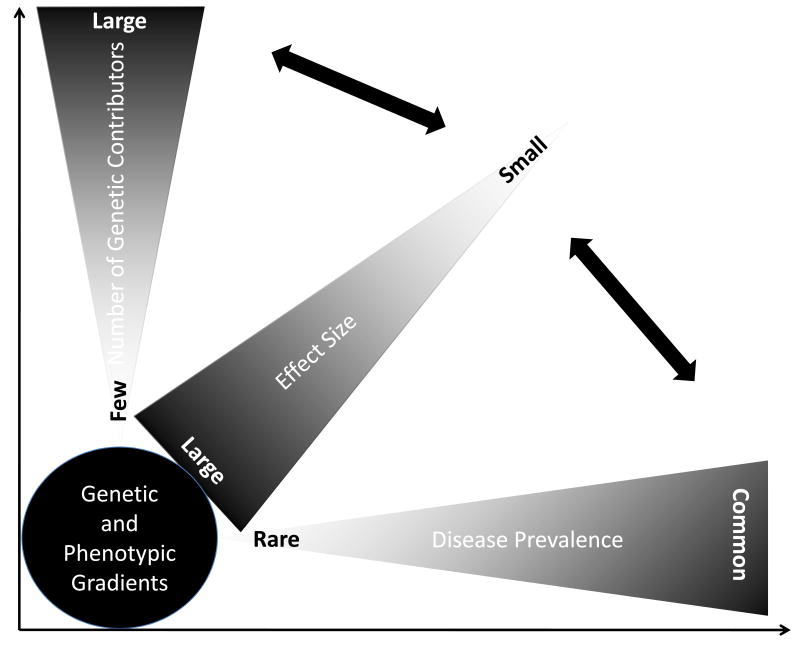
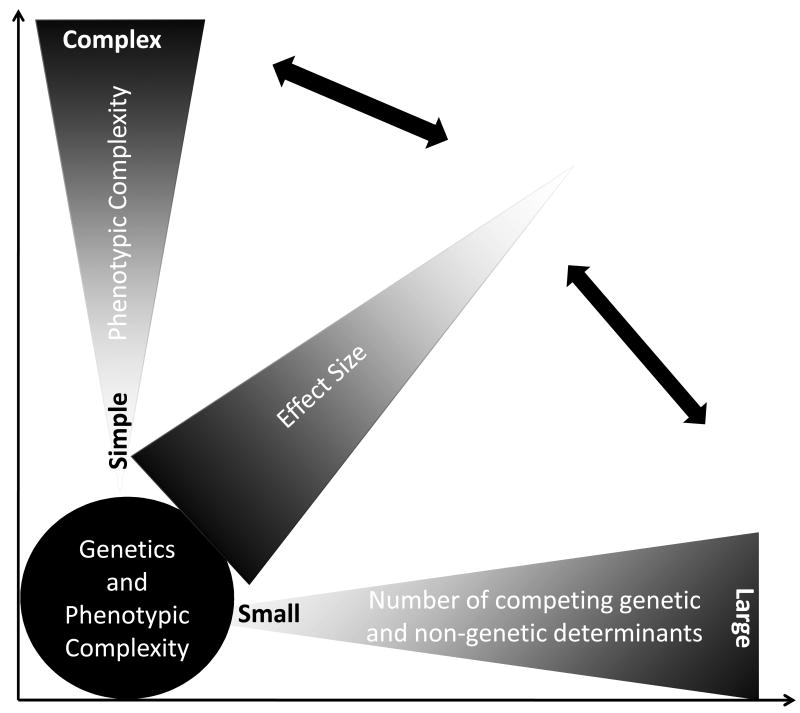
The effect sizes of DSVs on the expression of the phenotype follow a gradient ranging from negligible to profound [12]. In single gene disorders, the effect sizes of the DSVs are large. Consequently, the inheritance of such DSVs is necessary and sufficient to cause the disease, albeit the phenotypic expression of the disease typically varies. In contrast, in the absence of the inheritance of such DSVs the family member is at exceedingly low risk of the phenotype. Variability in the phenotypic expression of single gene disorders is in part due to the presence of additional DSVs that impart various effect sizes on the phenotype as well as other genetic and non-genetic factors. The number of DSVs in the human genome that impart large effect sizes on the clinical phenotype is relatively small as one may deduce from the prevalence of the single gene diseases (Figure 1). In contrast, the vast majority of the DSVs in the human genome impart modest and often indiscernible effect sizes on the clinical phenotype. This is typically the case for common cardiovascular diseases, such as atherosclerosis or hypertension, wherein a few, if any, DSVs impart more than modest effect sizes. Typically, thousands or more DSVs contribute to the phenotype, each imparting practically imperceptible effect. Overall, the gradient of effect sizes of the DSVs in the genome inversely mirrors the population frequencies of the variants (Figure 1). DSVs that impart major effect sizes are infrequent in the population, while those with modest effect sizes are more common. In addition, the spectrum of the gradient of effect sizes increases as the complexity of the phenotype increases. The more remote is the phenotype from the genotype, the smaller is the effect size of each DSV, because of the dilution of its effect size by the other DSVs and non-genetic factors, and the greater the number of contributing DSVs (Figure 2). In accord with this concept, one would expect a stronger effect of the DSVs on proximal phenotypes, such as the mRNA and protein levels than on distant phenotype, such as the clinical outcomes and death. The shortcomings of the DSVs in predicting the clinical phenotype is illustrated in the results of Genome-Wide Association Studies (GWAS), wherein the identified DSVs account only for a small fraction of the variability of the clinical phenotypes. Larger GWAS would be expected to further support the notion that a very large number of DSVs contribute to the phenotypic expression of complex clinical phenotypes, each exerting an infinitesimal effect. For example, it is estimated that about 93,000 SNPs account for 80% of the inter-individual variation in height, which is predominantly a heritable trait [13].
Figure 1. Prevalence of the disease, frequencies of the DNA sequence variants and the effect sizes.
In rare and typically single gene diseases, a single DNA sequence variant imparts a large effect size and a few others contribute to the phenotype as modifiers of the phenotype. In contrast, in common complex disorders a large number of DNA sequence variants contribute to the phenotype, each imparting modest and often clinically indiscernible effect sizes.
Figure 2. Phenotypic complexity and contributions of genetic and non-genetic determinants.
The more complex the phenotype is, such as the clinical outcomes, the greater the number of contributing factors to the phenotype and the smaller the effect sizes of each DNA sequence variant on the phenotype.
Linking the DNA sequence variations in the human genome to the clinical phenotype is the most challenging component of the “personalized Medicine” [14]. The clinical outcome is a complex phenotype that is not solely determined by the DSVs. It typically results from intertwined, stochastic, non-linear, dynamic ands often context-dependent interactions among the various contributors to the phenotype, whether genetic or non-genetic. There is no question that the genome is a very important determinant of the phenotype. At minimum, it provides the stage on which various constituents choreograph expression of the clinical phenotype. The genome, however, encompasses a multi-layer complexity that often is not evident from the analysis of its sequence variants. For example, the contributions of CNVs, intronic sequence variants, non-coding RNAs, such as microRNAs [15], alternatively spliced mRNA species, which relates 94% of the human genes [16] and the epigenetics [17] to the phenotypic expression of clinical phenotypes are poorly understood. Likewise, the impacts of post-translational modifications of proteins, such as phosphorylation, farnesylation and ubiquitinylation are expected to influence the expression of the clinical phenotype. Thus, to fully understand the genetic and non-genetic determinants of the clinical phenotype, it is essential to decipher all constituents that contribute to the clinical phenotype and incorporate them into the modeling, while realizing that no modeling is perfect. Accordingly, the enormous complexities of the human genome and the clinical phenotype expose the limitations of the diagnostic utility of DSVs, particularly for the common cardiovascular disorders. The clinical utility of the vast majority of the DSVs in the genome as diagnostic and prognostic tools for complex cardiovascular phenotype is expected to fall short of fulfilling Dr. Koshland's “Cha-Cha-Cha” theory of scientific discoveries [18]. Perhaps, a small number of such DSVs may offer limited diagnostic or prognostic values for complex phenotype but it remains to be proven. Nevertheless, in the gradient of phenotypic simplicity to complexity, with the decreasing complexity of the phenotype the potential clinical utility of DSVs increases. Thus, in contrast to common complex phenotypes, one might be able to extract valuable diagnostic and prognostic information from the DSVs that exert large effect sizes under certain clinical circumstances.
Conclusions
On the whole, the primary utility of DSVs in “personalized medicine” is in providing clues to the molecular mechanisms that govern the pathogenesis of the human diseases, whether complex or single gene diseases, and then applying the mechanistic knowledge to the cure of human diseases. Such discovery are likely to fulfill the Dr. Koshland's “Cha-Cha-Cha” theory of scientific discoveries [18].
Acknowledgments
Supported by grants from the NHLBI, Clinical Scientist Award in Translational Research from the Burroughs Wellcome Fund and The TexGen Fund from the Greater Houston Community Foundation
References
- 1.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Levy S, Sutton G, Ng PC, et al. The Diploid Genome Sequence of an Individual Human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** The first reporting of sequence of a diploid genome. The findings were truly remarkable as they showed that Dr. Venter sequence had about 4 million DNA sequence variations including more than 3 million single nucleotide polymorphisms and a very large number of structural variations. Subsequent publications of the sequence of genomes of other individuals have largely confirmed these findings.
- 4.Drmanac R, Drmanac S, Strezoska Z, et al. DNA sequence determination by hybridization: a strategy for efficient large-scale sequencing. Science. 1993;260:1649–52. doi: 10.1126/science.8503011. [DOI] [PubMed] [Google Scholar]; * The manuscript reports the sequence of a human genome by massively parallel sequencing under $ 5,000.00 by a privately company.
- 5.Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–6. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This manuscript reports using the exon-capture technology and the Next Generation DNA Sequencing to sequence all exons (exoms) in 12 genomes. They also report a proof-of principle discovery of a causative mutation by this approach.
- 6.Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–9. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheeler DA, Srinivasan M, Egholm M, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 8.Kim JI, Ju YS, Park H, et al. A highly annotated whole-genome sequence of a Korean individual. Nature. 2009;460:1011–5. doi: 10.1038/nature08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Wang W, Li R, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–5. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korbel JO, Urban AE, Affourtit JP, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marian AJ. Nature's genetic gradients and the clinical phenotype. Circ Cardiovasc Genet. 2009;2:537–9. doi: 10.1161/CIRCGENETICS.109.921940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–8. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 14.Marian AJ. Clinical implications of the “personal” genome. Curr Atheroscler Rep. 2008;10:361–3. doi: 10.1007/s11883-008-0055-6. [DOI] [PubMed] [Google Scholar]
- 15.Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol. 2009;21:461–9. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]; * The authors report on the enormous diversity of alternative splicing in the human genome. Accordingly, 94% of the genes in the human genome undergo some degree of alternative splicing.
- 17.Kaneda R, Takada S, Yamashita Y, et al. Genome-wide histone methylation profile for heart failure. Genes Cells. 2009;14:69–77. doi: 10.1111/j.1365-2443.2008.01252.x. [DOI] [PubMed] [Google Scholar]
- 18.Koshland Philosophy of Science: The Cha-Cha-Cha Theory of Scientific Discovery. Science. 2007;317:761–762. doi: 10.1126/science.1147166. [DOI] [PubMed] [Google Scholar]