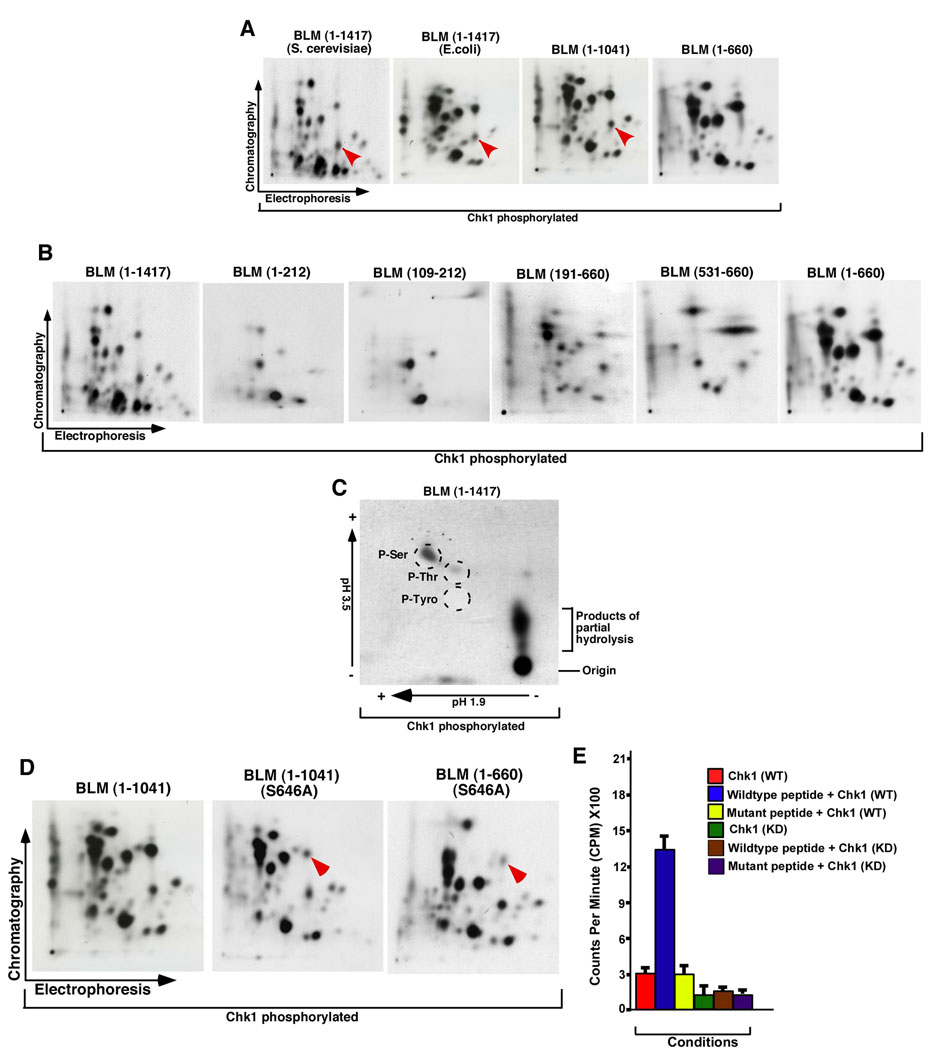
Figure 3. Chk1 phosphorylated BLM at Ser646 in vitro.
A. Phosphopeptide maps of human BLM (1–1417) (produced either in S. cerevisiae or in E. coli) and BLM fragments (1–1041) and (1–660) phosphorylated in vitro by Chk1. Red arrow indicates a phosphopeptide present in all except BLM (1–660). The black arrows indicate the directions in which the phosphopeptides were separated by electrophoresis and chromatography in the first and second dimensions, respectively.
B. Same as (A) except the following BLM derivatives were used: BLM (1–212), BLM (109–212), BLM (191–660), BLM (531–660), BLM (1–660) and BLM (1–1417).
C. Phosphoaminoacid analysis of BLM (1417). The arrows indicate the directions during the chromatographic runs. The broken circles indicate the positions of co-migrating cold phosphoaminoacid standards. The position of the origin and products of the partial hydrolysis have been indicated.
D. Phosphopeptide analysis of BLM (1–1041) and the two mutants BLM (1–1041) S646A and BLM (1–660) S646A. Arrow indicates the position of the phosphopeptide decreased in the mutants.
E. Peptides (containing either wild type or mutant Ser646 residue) were phosphorylated with either the wild type or kinase dead recombinant Chk1. Bound radioactivity was quantitated by scintillation counting.