Figure 3.
Factors Determining Cleavage Site Position
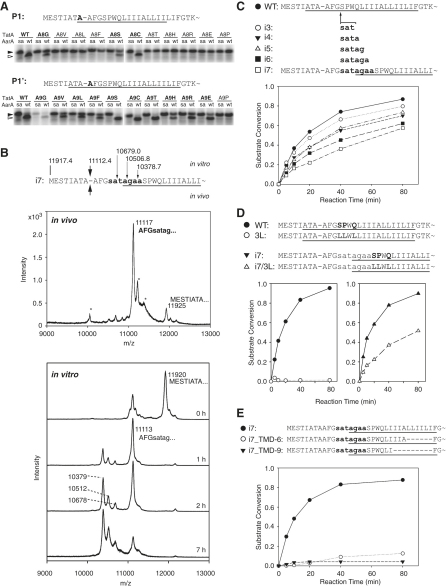
(A) Preferences at P1 and P1′ positions in TatA. A8 and A9 were mutated individually into amino acids whose side chains represent a range of physicochemical properties and analyzed for cleavage by AarA. The P1 position is much more constrained than P1′: tolerated mutations are highlighted in boldface. Enzyme concentration was 450 nM and reaction time 40 min. WT, wild-type; SA, catalytic serine-to-alanine mutant.
(B) A linker containing susceptible P1-P1′ pairs was inserted between G11 and S12 of TatA to generate the i7 mutant. As indicated by the large arrows, it is cleaved by AarA in vivo only at the original A8-A9 site. Cleavage also occurs at the same site in vitro, and when overdigested, less-efficient secondary cleavages occur in the linker region (small arrows). The theoretical masses of the C-terminal fragments resulting from i7 cleavage at indicated sites are annotated above its sequence. Upper graph, MALDI mass spectrum of the in vivo-processed i7 mutant that had been expressed in wild-type P. stuartii and isolated from the membrane fraction. Experimental masses of [M+H] ions and corresponding N termini are indicated. The N-terminal sequence determined by Edman degradation is highlighted in bold. Masses of the minor peaks marked with asterisks could not be matched to any cleavage product, and their identity was not established. Lower graph, MALDI mass spectra of an in vitro cleavage reaction time course of the i7 mutant that had been expressed in E. coli ΔglpG and isolated from the membrane fraction. AarA concentration was 11.2 μM, and recombinant i7 was at 20 μM.
(C) Cleavage rate of linker insertion mutants in vitro is negatively proportional to linker length. AarA was at 280 nM.
(D) TM helix-destabilizing residues are less important when cleavage occurs outside the bilayer. Substitution of S12, P13, and Q14 by leucine completely blocks cleavage in vitro in the context of otherwise wild-type TatA, but not in the context of the i7 mutant. To ensure comparable cleavage rates between the pairs, AarA concentration was 280 nM for WT and 3L, and 840 nM for i7 and i7/3L.
(E) Cleavage rate comparison of TMD deletion variants of the i7 linker insertion mutant showed that even when cleavage occurs outside the TMD, the hydrophobic part of i7 TMD is still required. AarA concentration was 840 nM. In (C)–(E), within each experiment the in vitro-translated radiolabeled substrate variants were equimolar as judged by equal intensity of their bands on the autoradiograms, and the cleavage rates were quantitated as in Figure 1B.