Abstract
Holoprosencephaly (HPE), the most common malformation of the human forebrain, may arise due to interacting genetic and environmental factors. To date, at least 12 contributory genes have been identified. Fibroblast growth factor 8 (Fgf8) belongs to the FGF family of genes expressed in several developmental signaling centers, including the anterior neural ridge, which is implicated in midline anomalies in mice. In humans, FGF8 mutations have been previously reported in facial clefting and in hypogonadotropic hypogonadism, but have not been reported in patients with HPE. We screened 360 probands with HPE for sequence variations in FGF8 using High Resolution DNA Melting (HRM) and sequenced all identified variations. Here we describe a total of 8 sequence variations in HPE patients, including a putative loss-of-function mutation in 3 members of a family with variable forms of classic HPE, and relate these findings to the phenotypes seen in other conditions.
Key Words: Anterior pituitary, FGF8, Holoprosencephaly, HPE, IHH
Holoprosencephaly is the most common congenital malformation of the human forebrain, with a prevalence of 1 in 250 conceptions and 1 in 10,000–20,000 live births [reviewed in Muenke and Beachy, 2000; Dubourg et al., 2007; Monuki, 2007; Roessler and Muenke, 2010]. HPE results from incomplete division of the forebrain early in gestation and is exquisitely sensitive to both genetic and environmental insults during this critical period. HPE is traditionally categorized by the degree of anatomical malformations, from most severe to least severe, into alobar, semilobar, lobar, and middle interhemispheric variant types [Muenke and Beachy, 2000; Cohen, 2006; Dubourg et al., 2007; Solomon et al., 2010]. Facial features in patients with HPE range from the most severe form with cyclopia (a single eye), proboscis (a nose-like structure above a single eye), and cleft lip and palate, to less severe microforms including single maxillary central incisor (SMCI), microcephaly, and hypotelorism (closely spaced eyes) but with structurally normal brains by conventional neuroimaging [Muenke and Beachy, 2000; Cohen, 2006]. Severe neurological impairment is found in virtually all patients with brain malformations [Muenke and Beachy, 2000; Dubourg et al., 2007; Lacbawan et al., 2009].
Although all potential causes of HPE are not well characterized, there is mounting evidence showing that interacting environmental and genetic factors are involved, and that severe manifestations may be due to a complex interaction of etiologies [Ming and Muenke, 2002; Bendavid et al., 2006; Dubourg et al., 2007; Roessler and Muenke, 2010].
Human FGF8 is a member of the large fibroblast growth factor (FGF) family and maps to chromosomal region 10q24 [Lorenzi et al., 1995; White et al., 1995; Payson et al., 1996; Yoshiura et al., 1997]. Fgf8 contains 6 exons and alternative splicing can result in 4 possible coding regions for the amino-terminus [Gemel et al., 1996; Olsen et al., 2006]. Vertebrate Fgf8 is involved in a variety of activities including mitotic induction and cell survival, and has been implicated in a broad range of biological processes, including cell growth, tissue repair, and tumor growth invasion [Yoshiura et al., 1997]. The temporal and spatial expression pattern of Fgf8 suggests its key involvement in embryonic development. In support of this, animal models show involvement of Fgf8 in limb development, gastrulation, left-right axis determination, and organ morphogenesis [Ohuchi et al., 1994; Johnson and Tabin, 1997; Meyers and Martin, 1999].
Recent studies have shed light onto the conserved vertebrate process by which secreted factors produced in patterning centers regulate early organ formation such as the developing forebrain [Rubenstein et al., 1998; Storm et al., 2006; Monuki, 2007; Fernandes and Hébert, 2008]. Specifically, in telencephalic specification multiple patterning centers contribute to the development of the midline of the forebrain, an axis essential for the division of the cerebral hemispheres that fails in the HPE sequence. Communication between these centers coordinates telencephalic development [Monuki, 2007; Fernandes and Hébert, 2008]. Through these patterning centers, Fgf8 signaling affects the telencephalon at several levels. Fgf8 disruption in both mouse and zebrafish results in a small telencephalon [Meyers et al., 1998]. The rostral patterning center expresses multiple Fgf genes, including Fgf8 [Maruoka et al., 1998]. Fgf8 expression in the rostral patterning center aids proliferation and inhibits differentiation and dorsal telencephalic fates [Xuan et al., 1995]. The dorsal patterning center, a center known to express Zic2 (mutations in ZIC2 cause HPE in humans), also expresses Bmp and Wnt genes, and directs development of cortical hem and dorsocaudal structures [Brown et al., 1998; Grove et al., 1998; Galceran et al., 2000]. Finally, the ventral patterning center expresses Shh (again, mutations in which cause HPE in humans) [Crossley and Martin, 1995; Shimamura et al., 1995; Roessler et al., 1996, 2009]. Shh interacts with Fgf8, and regulation of Shh expression and development of ventral telencephalon is known to be dependent on FGF signaling in zebrafish and other vertebrates [Shinya et al., 2001; Garel et al., 2003; Lupo et al., 2006; Storm et al., 2006; reviewed in Hoch et al., 2009]. These observations provide evidence for cross-linked communication between patterning centers and for the obligatory role of the FGF8 in communication between ventral and dorsal centers. Therefore, the testable prediction would be to measure the actual presence or absence of FGF8 variants among our HPE cases.
While mutations in FGF8 have not previously been studied in human HPE, heterozygous mutations in FGF8 can be found in other diseases in humans [reviewed in Krejci et al., 2009]. Mutations in FGF8 were found in 6 unrelated patients with idiopathic hypogonadotropic hypogonadism (IHH) with and without anosmia [Falardeau et al., 2008; for a review, see Bianco and Kaiser, 2009]. Functional studies show that all of these mutations resulted in diminished function of various degrees. Interestingly, Fgf8 hypomorphic mice show olfactory bulb anomalies, with reduced neuronal gonadotropin cells in the hypothalamus. Of note, one of the above patients with adult-onset IHH has an identical mutation to the one we found in HPE patients [Falardeau et al., 2008; see below].
Additionally, an FGF8 mutation (whose likely pathogenecity was inferred by structural analysis) has also been reported in one patient with nonsyndromic cleft lip and palate [Riley et al., 2007]. Importantly, all patients described so far with FGF8 mutations have phenotypes consistent with midline defects [Krejci et al., 2009].
As Fgf8 appears to act in midline induction by serving as an intermediate factor in a cross-regulatory interaction between ventral and dorsal domains, we felt that FGF8 was an excellent candidate gene for human HPE [Storm et al., 2006; Monuki, 2007; Fernandes and Hébert, 2008; Paek and Hébert, 2009]. Here we will present the use of high-resolution DNA melting (HRM), with bi-directional sequencing of variants as a robust and sensitive screening methodology to evaluate new HPE candidate genes with respect to their potential roles as primary or secondary factors in forebrain development pathologies.
Materials and Methods
The genomic organization of the FGF8 gene (NM_033164.1, corresponding to the longest transcript: variant F) was obtained from public database programs (http://www.ncbi.nlm.nih.gov) and the Bioinformatics site annotation provided by http://www.genome.ucsc.edu. Guidelines for the naming of the sequence variants conform to the recommendations of the human nomenclature committee (www.hgvs.org/mutnomen). Oligonucleotide primers were designed using the Primer3 v. 0.4.0 and Oligo™ 6.8 for the 6 coding exons and exon/intron boundaries of all known isoforms, as described (table 1).
Table 1.
Primers for PCR amplification of the human FGF8 gene
| Exon | Forward | Reverse | Product size, bp | Tm, °Ca |
|---|---|---|---|---|
| 1 | 5′-AGCGGCGCACAGCGATTCGGTG-3′ | 5′-G AGGCAG AGGACCAGCAAGTGC-3′ | 343 | 66.4 |
| 2 | 5′-CCGCTTTTGTCTCCCACAG-3′ | 5′-G ACCTCAGGAGGGGTGCTAC-3′ | 245 | 56.4 |
| 3 | 5′-CCAGGGGATGGATGTTCG-3′ | 5′-CCACCTGTTGGGAGACAC-3′ | 303 | 64 |
| 4 | 5′-CCTGGGCGGAGTAGCATTA-3′ | 5′-TCCCACAAGCTACCTTCAGC-3′ | 264 | 65 |
| 5 | 5′-GCCTTCTGCCTACCTTGTTG-3′ | 5′-ATCTGCCAAT AGCCATCCTG-3′ | 254 | 57 |
| 6.1 | 5′-GGGTGCCCTACAGGATGAG-3′ | 5′-ACCAGCGTGAGGTCCACTT-3′ | 206 | 63.5 |
| 6.2 | 5′-GGGCGGGTAGTTGAGGAAC-3′ | 5′-ACGG AGATTGTGCTGGAGA-3′ | 207 | 62.4 |
| 6.3 | 5′-GGTGAAGGCCATGTACCAG-3′ | 5′-GCGAGTTGTG AGGGATTAGAG A-3′ | 270 | 64.2 |
Annealing temperature for PCR amplification.
Screened Populations
Our study consists of 360 unrelated cases from our collection of probands with HPE, which encompasses the entire clinical spectrum of severity in our HPE patients. In addition, our study also includes 184 unrelated individuals acquired as de-identified samples from the Coriell Institute for Medical Research (Camden, N.J.) to serve as controls. High-resolution melting profile was performed on the 360 unrelated HPE patients and the 184 unrelated controls simultaneously. Investigation of all samples was in accordance with a National Human Genome Research Institute (NHGRI) Institutional Review Board-approved research protocol.
PCR Amplification
Roche LightCycler® 480: All exons, except exon 1 (see below), were amplified in a 7.5-μl reaction volume using 5–10 ng DNA template, 3.5 μl of Roche High-resolution melting Master (containing FastStart Taq DNA polymerase, reaction buffer, dNTP mix, and HRM dye (Roche)), 0.6 μl of magnesium chloride (Roche) and 0.3 mM of each primer. All reactions were done employing a Roche LightCycler® 480 (Roche, Ind.). PCR amplification parameters were: incubation at 95°C for 10 min followed by 45 cycles of denaturation at 95°C for 10 s, annealing (depending on primer, see table 1) for 15 s, and extension at 72°C at amplicon size divided for 25 s.
PCR Assay for Exon 1: Exon 1 was amplified using a 25-μl reaction volume using 5–10 ng DNA template, 0.2 mM of dNTP, 0.75 mM of each primer, 2.5 μl of 10× PCR buffer (Invitrogen), 2.5 μl of 10× Enhancer buffer (Invitrogen), 0.75 μl of magnesium sulfate (Invitrogen) and 0.2 μl Taq polymerase. Every reaction was performed using a PTC-225 thermocycler (MJ Research, Mass.). PCR cycling parameters were: 95°C for 4 min followed by 40 cycles at 95°C, annealing at 66.4°C, extension at 72°C for 30 s, and a final extension step of 72°C for 7 min.
High-Resolution DNA Melting Acquisition
High-resolution melting curve analysis was used to identify putative sequence variants in FGF8 in 360 HPE samples and 184 normal controls using the LightCyler® 480 II system (Roche applied science, Indianapolis, Ind.). Samples in all exons, except exon 1, were melted by increasing the temperature from 65 to 95°C, while exon 1 was melted from 65 to 99°C at a programmed rate of 0.2°C/s with fluorescence acquisition of 25 times per 1°C of temperature increase.
High-Resolution DNA Melting Analysis
384 well DNA-containing sample plates were used to scan for mutations; within these plates, 16 water-containing blank samples were used as negative controls. HRM curve analysis was performed according to the manufacturer's recommendations using LightCycler® 480 Software release v.1.5.0 (Idaho Technology Inc., Idaho and Roche Applied Sciences, Ind.). Amplicons of individuals showing evidence for variants were normalized, temperature-shifted fluorescence-over-temperature plots were identified, and samples were sequenced bi-directionally to confirm the presence of sequence changes.
DNA Sequencing
Samples with melting profiles deviating from the profile of the wild type were sequenced to define the variant. Sequencing was performed bi-directionally at the core DNA Sequencing Facility, National Institute of Neurological Disorders and Stroke, NIH.
Clinical Data
In accordance with our NHGRI IRB-approved procedures, families with positive clinical or molecular findings were contacted and consented to participate in our comprehensive clinical study on HPE.
Results
Molecular Findings in HPE Patients and Controls
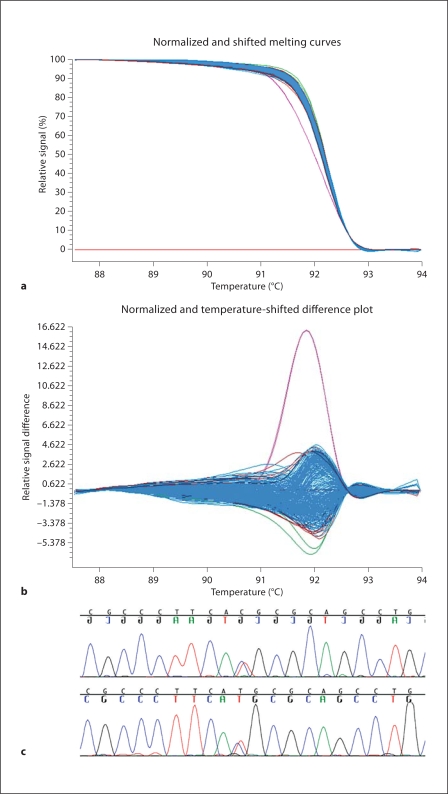
We find both novel and known variants in FGF8 in both HPE patients and controls (table 2). Most of these variations occur in non-coding intronic or flanking sequences that do not have any known functional effects. In 1 HPE-affected family, we find a hypomorphic mutation in a highly evolutionary conserved region of FGF8: c.686C>T, resulting in p.T229M (fig. 1). This mutation results in the substitution of a hydrophobic amino acid (methionine) for a hydrophilic amino acid (threonine) in a region of the molecule present in all 4 known biologically active isoforms. Of note, this specific mutation has been reported in a patient with idiopathic hypogonadotropic hypogonadism, was shown to have a deleterious functional effect using in vitro models, and is predicted to alter activity of FGF8 in humans [Falardeau et al., 2008]. While the precise mechanism underlying its reduced activity is unknown, due to its location in the extreme carboxy-terminus of the protein, it has been recently demonstrated that factors other than direct binding to receptors likely contribute to sharp morphogenetic gradients in biological tissues [Yu et al., 2009]. By bidirectional sequencing, no coding region mutations were found in the four most common HPE-associated genes in this family (SHH, ZIC2, SIX3, and TGIF).
Table 2.
Variants detected in FGF8 by HRM and bi-directional sequencing. DNA mutation numbering is based on the cDNA sequence. A of ATG translation initiation codon is +1 and the initiation codon is codon 1 based on NM_033163.1 (for a review, see Krejci et al. [2009])
| Exon | Nucleotide and amino acid variants | Location | Number of HPE patients | Number of controls |
|---|---|---|---|---|
| 1 | c.l-35C>T [5′UTR-35C>T, rs2735432] | 5′UTR | 8 | 3 |
| 1 | c.l-6G>T [5′UTR-6G>T] | 5′UTR | 1 | 0 |
| 3 | c.70-127G>C [IVS2-127G>C] | intronic | 1 | 0 |
| 4 | c.232C>T [p.R78C] | missense | 0 | 1 |
| 5 | c.444+19G>A [IVS5+19G>A, rs3218234] | intronic | 12 | 4 |
| 5 | c.444+30C>A [IVS5+30C>A] | intronic | 1 | 0 |
| 5 | c.444+36C>A [IVS5+36C>A] | intronic | 1 | 0 |
| 6 | c.678G>T [p.P226P] | synonymous substitution | 1 | 0 |
| 6 | c.686C>T [p.T229M] | missense | 2 | 0 |
Fig. 1.
HRM detection of variant c.686C>T, resulting in p.T229M in exon 6 of FGF8. a Temperature melt profile of the variant (in magenta) is distinct from wild types. b Differential plot illustrates the variant (in magenta) is distinct from wild types. c Representative chromatogram shows the DNA sequence variation detected in all 3 members of the family.
Clinical Findings
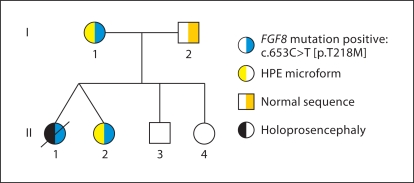
Three members of the family (fig. 2) tested positive for a mutation in FGF8: c.686C>T, resulting in p.T229M. This maternally-inherited mutation was initially found in dizygotic twin females. The first twin (II.1) had semilobar HPE, and features consistent with classic non-syndromic HPE, including microcephaly, cleft palate, seizures, diabetes insipidus (DI), and severe neurological impairment. Other than the presence of DI, she had no evidence for other endocrinological disturbances. The other twin (II.2) had an SMCI and hypotelorism. Brain MRI performed at approximately 1 year of age was described as showing subtle midline anomalies, with the presence of bilateral olfactory bulb dysplasia, but a repeat MRI performed at 8 years of age at the NIH showed no evidence of midline abnormalities. Significantly, this child was quite intellectually gifted, supported by standardized scholastic evaluations showing that she performed several years above her chronological age in reading and mathematics. The only clinical manifestation in the twins' mother (I.1) was mild hypotelorism; she was also highly intelligent by history. Her brain MRI showed no midline or other anomalies.
Fig. 2.
Pedigree of family segregating FGF8 mutation. As the legend indicates, individuals with the right side shaded blue segregate the c.686C>T mutation in FGF8. Individuals I.1 and II.2 display microform features, indicated by yellow shading, while patient II.1 had semilobar HPE, indicated by black shading. Individuals II.3 and II.4 were not tested, but by history and photo review had no findings consistent with microform HPE or other related issues.
Neither living family member with the mutation showed any signs of endocrinological disturbance despite a comprehensive evaluation. Specifically, there were no signs of hypogonadotropic hypogonadism by history, serum screening, or by radiologic anatomical study.
Discussion
We tested 360 individuals with HPE for mutations in FGF8. Surprisingly, we detected only 1 family with a putative loss-of-function mutation in this gene. The low incidence of mutations of the FGF8-coding region argues against mutations in this gene as a common genetic factor to explain the occurrence of midline forebrain anomalies. However, we cannot exclude the possibility that unscreened genetic elements could play a role in human disease, for example, those that control the levels or tissue distribution of FGF8 transcripts [Komisarczuk et al., 2009], or its various isoforms, which were not the focus of this study. Nor can we exclude the ability of other FGFs to compensate for any putative diminished FGF8 function.
Several additional conclusions can be drawn from this study. First of all, this family simultaneously highlights the importance and limitation of functional studies, both from the perspective of understanding the genetic pathogenesis of HPE, and in terms of counseling families in the clinical setting. The p.T229M mutation found in this family and in an apparently unrelated family with IHH occurs in an evolutionarily conserved region of FGF8 suggesting its biological relevance. Evidence from animal models and cell-based assays shows that this mutation has a modest deleterious functional effect. Specifically, this heterozygous mutation is predicted to alter FGF8 activity in humans [Falardeau et al., 2008]. However, it is reasonable to infer that additional genetic or environmental factors, presently unknown, would be required to produce severe phenotypes. Interestingly, several families with IHH have been described that segregate more than one type of mutation for this condition, and, therefore, an oligogenic model has been proposed [Pitteloud et al., 2007; Cole et al., 2008] that resembles the ‘multiple hit’ model of HPE in many respects [Ming and Muenke, 2002].
The family reported here shows that the same mutation can result in highly variable clinical consequences; this mutation must therefore not solely determine phenotypic outcome. This highly variable intrafamilial expressivity is typical in other families with mutations in HPE-related genes [Roessler et al., 1996; Solomon et al., 2009]. This inter- and intrafamilial variable expressivity is typical for HPE and other complex traits including IHH [Pitteloud et al., 2007; Cole et al., 2008]. This argues that FGF8 may act as one of numerous factors in the model of the pathogenesis of HPE resulting from interacting genetic and environmental influences [Ming and Muenke, 2002; Roessler and Muenke, 2010].
Second, previous studies have shown that patients with mutations in FGF8 are associated with midline defects such as cleft lip and palate and central pituitary/hypothalamic dysfunction. Using our cohort of patients with HPE, we show that a mutation in FGF8 may contribute to a midline defect at the most severe end of the phenotypic spectrum (table 3). However, we cannot rule out the possibility that other genetic and/or environmental factors are equally relevant to the pathologies seen in this family.
Table 3.
Putative disease-related FGF8 variants
| Patients | Nucleotide change | Amino acid substitution | Diagnosis | Associated phenotypes | Reference |
|---|---|---|---|---|---|
| 1a | c.686C>T | p.T229M | HPE | semilobar HPE, cleft palate, and seizures | this report |
| 1b | c.686C>T | p.T229M | microform HPE | microform HPE | this report |
| 1c | c.686C>T | p.T229M | microform HPE | very subtle microform HPE | this report |
| 2 | c.686C>T | p.T229M | AHH | type II diabetes | Falardeau et al. [2008] |
| 3 | c.40C>A | p.H14N | nIHH | high arched palate, osteoporosis, fractures | Falardeau et al. [2008] |
| 4 | c.77C>T | p.P26L | KS | none | Falardeau et al. [2008] |
| 5 | c.118T>C | p.F40L | nIHH | none | Falardeau et al. [2008] |
| 6 | c.298A>G | p.K100E | nIHH | microphallus | Falardeau et al. [2008] |
| 7 | c.379C>G | p.R127G | KS | cleft lip and palate, osteoporosis, hearing loss, hypotelorism, flat nasal bridge, camplodactyly, hyperlaxity | Falardeau et al. [2008] |
| 8 | c.217G>C | D73H | NS CLP | BL CLP | Riley et al. [2007] |
Abbreviations: AHH, adult-onset hypogonadotropic hypogonadism; BL, bilateral; KS, Kallmann syndrome; nIHH, normosmic idiopathic hypogonadotropic hypogonadism; NS CLP, nonsyndromic cleft lip and palate.
Our results suggest that mutations in an evolutionarily conserved region of FGF8 can affect human midline and forebrain development. This report, in conjunction with previous reports, provides evidence that FGF8 is involved in midline patterning in humans and that failure of its proper expression can infrequently result in human diseases. Despite evidence for the key role of FGF8 in the cascade involved in midline forebrain patterning, this study revealed a low prevalence of mutations in FGF8 in our cohort of patients with HPE. Biologically, FGF8 appears to play important roles in forebrain development, but may only act to contribute to frank disease in the presence of other, currently undetermined factors. Furthermore, the fact that the family described here did not show any signs of IHH indicates that other factors are almost certainly yet to be identified. Future directions should be centered on screening more patients with a variety of midline defects for mutations in FGF8 in order to determine the entire spectrum of phenotypic consequences. In conjunction, it will be important to continue to explore the presence of other contributory biological factors in these individuals.
Acknowledgements
We are extremely grateful to the family presented in this report and to all the other research participants. This research was supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health.
References
- Bendavid C, Haddad BR, Griffin A, Huizing M, Dubourg C, et al. Multicolour fish and quantitative PCR can detect submicroscopic deletions in holoprosencephaly patients with a normal karyotype. J Med Genet. 2006;43:496–500. doi: 10.1136/jmg.2005.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco SDC, Kaiser UB. The genetic and molecular basis of idiopathic hypogonadotropic hypogonadism. Nat Rev Endocrinol. 2009;5:569–576. doi: 10.1038/nrendo.2009.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Warburton D, Brown LY, Yu C, Roeder ER, et al. Holoprosencephaly due to mutations in ZIC2, a homologue of drosophila odd-paired. Nat Genet. 1998;20:180–183. doi: 10.1038/2484. [DOI] [PubMed] [Google Scholar]
- Cohen M. Holoprosencephaly: Clinical, anatomic, and molecular dimensions. Birth Defects Res A Clin Mol Teratol. 2006;76:658–673. doi: 10.1002/bdra.20295. [DOI] [PubMed] [Google Scholar]
- Cole LW, Sidis Y, Zhang C, Quinton R, Plummer L, et al. Mutations in Prokineticin 2 and Prokineticin receptor 2 genes in human gonadotrophin-releasing hormone deficiency: molecular genetics and clinical spectrum. J Clin Endocrinol Metab. 2008;93:3551–3559. doi: 10.1210/jc.2007-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- Dubourg C, Bendavid C, Pasquier L, Henry C, Odent S, David V. Holoprosencephaly. Orphanet J Rare Dis. 2007;2:8. doi: 10.1186/1750-1172-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falardeau J, Chung WCJ, Beenken A, Raivio T, Plummer L, et al. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J Clin Invest. 2008;118:2822–2831. doi: 10.1172/JCI34538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes M, Hébert JM. The ups and downs of holoprosencephaly: dorsal versus ventral patterning forces. Clin Genet. 2008;73:413–423. doi: 10.1111/j.1399-0004.2008.00994.x. [DOI] [PubMed] [Google Scholar]
- Galceran J, Miyashita-Lin EM, Devaney E, Rubenstein JLR, Grosschedl R. Hippocampus development and generation of dentate gyrus granule cells is regulated by LEF1. Development. 2000;127:469–482. doi: 10.1242/dev.127.3.469. [DOI] [PubMed] [Google Scholar]
- Garel S, Huffman KJ, Rubenstein JLR. Molecular regionalization of the neocortex is disrupted in Fgf8 hypomorphic mutants. Development. 2003;130:1903–1914. doi: 10.1242/dev.00416. [DOI] [PubMed] [Google Scholar]
- Gemel J, Gorry M, Ehrlich GD, MacArthur CA. Structure and sequence of human FGF8. Genomics. 1996;35:253–257. doi: 10.1006/geno.1996.0349. [DOI] [PubMed] [Google Scholar]
- Grove EA, Tole S, Limon J, Yip LW, Ragsdale CW. The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development. 1998;125:2315–2325. doi: 10.1242/dev.125.12.2315. [DOI] [PubMed] [Google Scholar]
- Hoch RV, Rubenstein JLR, Pleasure S. Genes and signaling events that establish regional patterning of the mammalian forebrain. Sem Cell Dev Biol. 2009;20:378–386. doi: 10.1016/j.semcdb.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Johnson RL, Tabin CJ. Molecular models for vertebrate limb development. Cell. 1997;90:979–990. doi: 10.1016/s0092-8674(00)80364-5. [DOI] [PubMed] [Google Scholar]
- Komisarczuk AZ, Kawakami K, Becker TS. Cis-regulation and chromosomal rearrangement of the fgf8 locus after the teleost/terapod split. Dev Biol. 2009;336:301–312. doi: 10.1016/j.ydbio.2009.09.029. [DOI] [PubMed] [Google Scholar]
- Krejci P, Prochazkova J, Bryja V, Kozubik A, Wilcox WR. Molecular pathology of the fibroblast growth factor family. Hum Mut. 2009;30:1245–1255. doi: 10.1002/humu.21067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacbawan F, Solomon BD, Roessler E, El-Jaick K, Domené S, et al. Clinical spectrum of SIX3-associated mutations in holoprosencephaly: correlation between genotype, phenotype and function. J Med Genet. 2009;46:389–398. doi: 10.1136/jmg.2008.063818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzi MV, Long JE, Miki T, Aaronson SA. Expression cloning, developmental and chromosomal localization of fibroblast growth factor-8. Oncogene. 1995;10:2051–2055. [PubMed] [Google Scholar]
- Lupo G, Harris WA, Lewis KE. Mechanisms of ventral patterning in the vertebrate nervous system. Nature Rev Neurosci. 2006;7:103–114. doi: 10.1038/nrn1843. [DOI] [PubMed] [Google Scholar]
- Maruoka Y, Ohbayashi N, Hoshikawa M, Itoh N, Hogan BLM, Furuta Y. Comparison of the expression of three highly related genes, Fgf8, Fgf17 and Fgf18, in the mouse embryo. Mech Dev. 1998;74:175–177. doi: 10.1016/s0925-4773(98)00061-6. [DOI] [PubMed] [Google Scholar]
- Meyers EN, Martin GR. Differences in left-right axis pathways in mouse and chick: Functions of FGF8 and SHH. Science. 1999;285:403–406. doi: 10.1126/science.285.5426.403. [DOI] [PubMed] [Google Scholar]
- Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- Ming JE, Muenke M. Multiple hits during early embryonic development: Digenic diseases and holoprosencephaly. Am J Hum Genet. 2002;17:1017–1032. doi: 10.1086/344412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monuki ES. The morphogen signaling network in forebrain development and holoprosencephaly. J Neuropathol Exp Neurol. 2007;66:566–575. doi: 10.1097/nen.0b013e3180986e1b. [DOI] [PubMed] [Google Scholar]
- Muenke M, Beachy PA. Genetics of ventral forebrain development and holoprosencephaly. Curr Opin Genet Dev. 2000;10:262–269. doi: 10.1016/s0959-437x(00)00084-8. [DOI] [PubMed] [Google Scholar]
- Ohuchi H, Yoshioka H, Tanaka A, Kawakami Y, Nohno T, Noji S. Involvement of androgen-induced growth factor (FGF-8) gene in mouse embryogenesis and morphogenesis. Biochem Biophys Res Commun. 1994;204:882–888. doi: 10.1006/bbrc.1994.2542. [DOI] [PubMed] [Google Scholar]
- Olsen SK, Li JY, Bromleigh C, Eliseenkova AV, Ibrahimi OA, et al. Structural basis by which alternative splicing modulates the oragnizer activity of FGF8 in the brain. Genes Dev. 2006;20:185–198. doi: 10.1101/gad.1365406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek H, Hébert JM. FGF signaling is strictly required to maintain early telencephalic precursor cell survival. Development. 2009;136:2457–2465. doi: 10.1242/dev.032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payson RA, Wu J, Liu Y, Chiu MI. The human FGF-8 localizes on chromosome 10q24 and is subjected to induction by androgen in breast cancer cells. Oncogene. 1996;1:47–53. [PubMed] [Google Scholar]
- Pitteloud N, Quinton R, Pearce S, Raivio T, Acierno J, et al. Digenic mutations account for variable penotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest. 2007;117:457–463. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BM, Mansilla MA, Ma J, Daack-Hirsch S, Maher BS, et al. Impaired FGF signaling contributes to cleft lip and palate. Proc Natl Acad Sci USA. 2007;104:4512–4517. doi: 10.1073/pnas.0607956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Muenke M. The molecular genetics of holoprosencephaly. Am J Med Genet C Semin Med Genet. 2010;154C:52–61. doi: 10.1002/ajmg.c.30236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Belloni E, Gudenz K, Jay P, Berta P, et al. Mutations in the human sonic hedgehog gene cause holoprosencephaly. Nature Genetics. 1996;14:357–360. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- Roessler E, El-Jaick KB, Dubourg C, Vélez JI, Solomon BD, et al. The mutational spectrum of holoprosencephaly-associated changes within the SHH gene in humans predicts loss-of-function through either key structural alterations of the ligand or its altered synthesis. Hum Mutat. 2009;30:E921–E935. doi: 10.1002/humu.21090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL, Shimamura K, Martinez S, Puelles L. Regionalization of the prosencephalic neural plate. Ann Rev Neurosci. 1998;21:445–477. doi: 10.1146/annurev.neuro.21.1.445. [DOI] [PubMed] [Google Scholar]
- Shimamura K, Hartigan DJ, Martinez S, Puelles L, Rubenstein JLR. Longitudinal organization of the anterior neural plate and neural tube. Development. 1995;121:3923–3933. doi: 10.1242/dev.121.12.3923. [DOI] [PubMed] [Google Scholar]
- Shinya M, Koshida S, Sawada A, Kuroiwa A, Takeda H. Fgf signalling through MAPK cascade is required for development of the subpallial telencephalon in zebrafish embryos. Development. 2001;128:4153–4164. doi: 10.1242/dev.128.21.4153. [DOI] [PubMed] [Google Scholar]
- Solomon BD, Lacbawan F, Jain M, Domené S, Roessler E, et al. A novel SIX3 mutation segregates with holoprosencephaly in a large family. Am J Med Genet A. 2009;149A:919–925. doi: 10.1002/ajmg.a.32813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon BD, Mercier S, Vélez JI, Pineda-Alvarez DE, Wyllie A, et al. Analysis of genotype-phenotype correlations in human holoprosencephaly. Am J Med Genet C Semin Med Genet. 2010;154C:133–141. doi: 10.1002/ajmg.c.30240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm EE, Garel S, Borello U, Hebert JM, Martinez S, et al. Dose-dependent functions of Fgf8 in regulating telencephalic patterning centers. Development. 2006;133:1831–1844. doi: 10.1242/dev.02324. [DOI] [PubMed] [Google Scholar]
- White RA, Dowler LL, Angeloni SV, Pasztor LM, MacArthur CA. Assignment of FGF8 to human chromosome 10q25–q26: mutations in FGF8 may be responsible for some types of acrocephalosyndactyly linked to this region. Genomics. 1995;30:109–111. doi: 10.1006/geno.1995.0020. [DOI] [PubMed] [Google Scholar]
- Xuan S, Baptista CA, Balas G, Tao W, Soares VC, Lai E. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron. 1995;14:1141–1152. doi: 10.1016/0896-6273(95)90262-7. [DOI] [PubMed] [Google Scholar]
- Yoshiura K, Leysens NJ, Chang J, Ward D, Murray JC, Muenke M. Genomic structure, sequence, and mapping of human FGF8 with no evidence for its role in craniosynostosis/limb defect syndromes. Am J Med Genet. 1997;72:354–362. [PubMed] [Google Scholar]
- Yu SR, Burkhardt M, Pies NM, Petrásek Z, Schloop S, Schwille P, Brand M. Fgf8 morphogen gradient forms by a source-sink mechanism with freely diffusing molecules. Nature. 2009;461:533–536. doi: 10.1038/nature08391. [DOI] [PubMed] [Google Scholar]