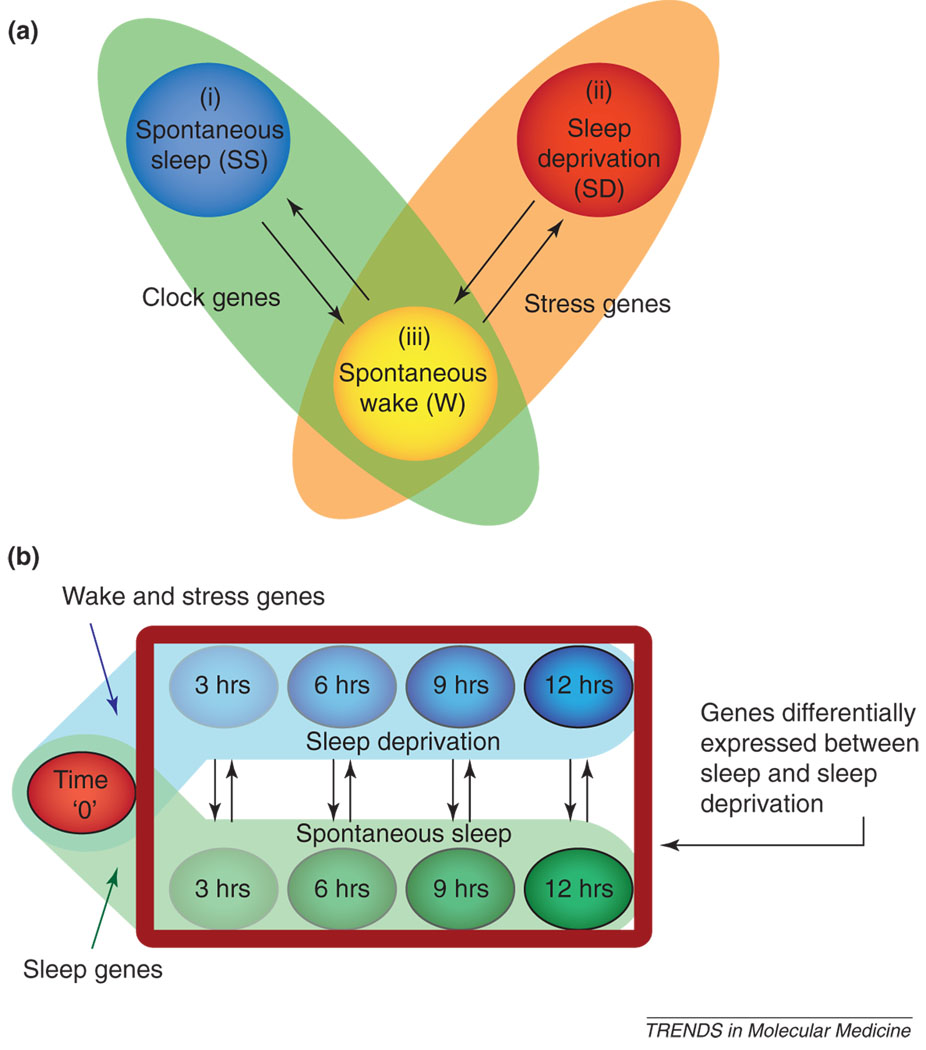
Figure 1.
Examples of experimental strategies in microarray research to elucidate the identity of genes whose expression exhibits differences between bouts of sleep and wakefulness. These complementary strategies address the confounding effects of sleep deprivation-induced systemic stress and the circadian clock on gene expression. (a) This approach [13] used three groups of rats: (i) a group of spontaneously sleeping (SS) animals who were required to sleep before sacrifice in the lights-on condition at least 75% of the previous 8 h; (ii) a group of sleep-deprived (SD) animals kept awake continuously by gentle handling and sacrificed at the same diurnal time as the SS group; and (iii) a group of animals that had a period of spontaneous wakefulness for 70% of the previous 8 h during the lights-out condition (W). Genes whose expression is higher during sleep, yet similar (lower) during sleep deprivation and the spontaneously awake condition (SS > SD = W) are called ‘sleep genes’, whereas genes conforming to the condition SS < SD = W are called ‘wake genes’. Although this design controls for stress and circadian confounders, it does not assess the direction of change in gene expression changes (i.e. up- or downregulation) or identify dynamic changes in transcriptional response. (b) This longitudinal study design [17] compares gene expression in animals allowed to sleep (green circles) before sacrifice with animals that are sleep deprived (blue circles) for different durations but sacrificed at the same time of day to control for circadian influences. An additional group of animals at ‘time zero’ (red circle) enables estimation of the direction of change for genes found to be expressed differentially between sleep and sleep deprivation. In this case, the experimental design controls for circadian influences in the estimation of temporal changes but does not take into account the effects of stress on gene expression in the group of animals subjected to sleep deprivation. Temporal changes in expression in the sleep groups are, however, not affected by stress because these animals are undisturbed. In both (a) and (b), black arrows indicate key comparisons; the strategy described in (a) relies on the fold-change, whereas the strategy in (b) relies on false-discovery rates as ‘filtering’ tools to identify differentially expressed genes. The false-discovery rate is a statistical procedure to control the number of mistakes made when performing multiple hypothesis tests, such as those performed during a search for differentially expressed genes in microarray experiments [102].