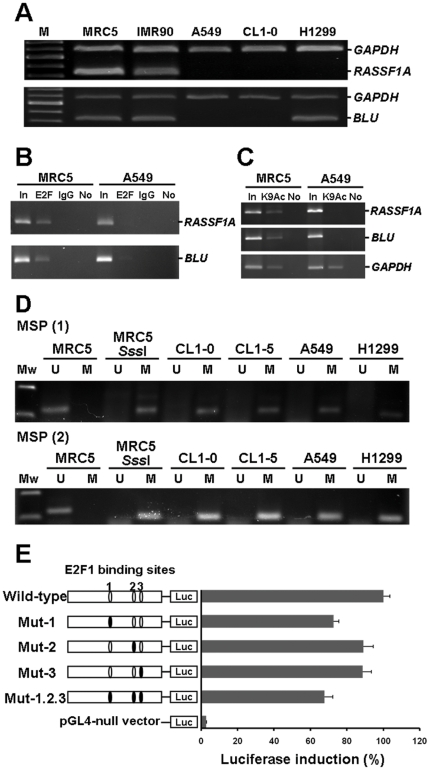
Figure 2. E2F1 bound to the RASSF1A promoter at potential transcriptionally important CpGs.
(A) RT-PCR analysis for RASSF1A and BLU gene expressions in MRC5 and IMR90 normal cells and three cancer cell lines (A549, CL1-0, and H1299). GAPDH was amplified as an internal control. (B) ChIP-PCR analysis for E2F1 binding to RASSF1A and BLU sequences in MRC5 normal cell and A549 cancer cell line. The binding of E2F1 to RASSF1A and BLU promoters was observed in MRC5 cell line. “In”, total input DNA; “E2F”, DNA-protein complex precipitated by anti-E2F1; “IgG”, DNA-protein complex precipitated by rabbit IgG; and “No”, no antibody. (C) ChIP assay with anti-acetylated histone H3 for RASSF1A, BLU and GAPDH loci. “K9Ac”, DNA-protein complex precipitated by anti-acetylated histone H3. (D) Methylation status of E2F1 binding sites in RASSF1A gene was assessed by MSP in MRC5 normal cell and four cancer cell lines. U: unmethylated gene; M: methylated gene. SssI methyltransferase-treated MRC5 DNA was used as methylation positive control. (E) Relative luciferase activities of different E2F1 mutated constructs are shown on the right side as bars in the bar chart, with the structure of each construct shown on the left side. Luc: luciferase gene sequence. E2F1-expression vector was cotransfected with different E2F1 binding sites mutated constructs (mutation sites shown as black circles).