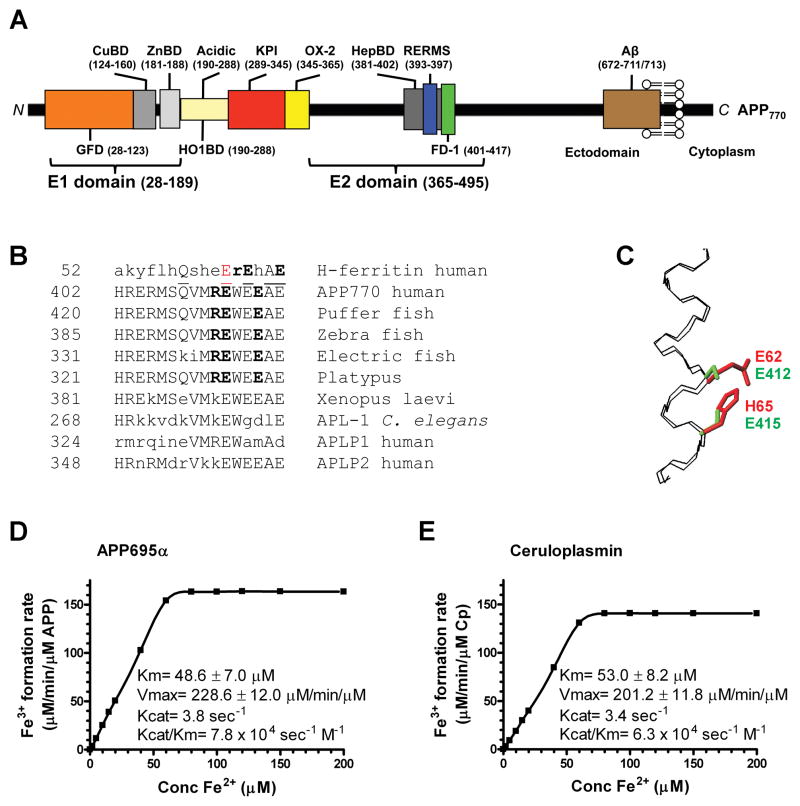
Figure 1. Characterization of APP695α ferroxidase activity.
A, Schematic of APP domains. The APP770 isoform is shown, APP751 lacks the OX-2 domain, and APP695 lacks both OX-2 and Kunitz protease inhibitor (KPI) domains. CuBD= copper binding domain, ZnBD= zinc binding domain. B, Sequence homologies for the REXXE motif. A sole match for the REXXE motif (in bold) of H-ferritin is at residues 411–415 of human APP770, commencing 5 residues downstream from the RERMS neurotrophic motif (Ninomiya et al., 1993). This is an evolutionarily-conserved motif not present in either human APLP1 or APLP2. A consensus alignment of three glutamate residues and the ferroxidase active site of H-ferritin is underlined. The first glutamate of the REWEE motif of APP could be aligned with Glu62 of H-ferritin (in red), which is part of the ferroxidase catalytic site (Lawson et al., 1989; Toussaint et al., 2007) although this forces the REXXE motifs of the proteins two residues out of register. C, An overlay of the backbone atoms (N, Cα, C) of residues 52–67 of the known H-ferritin active site (Lawson et al., 1991) (PDB accession no. 1FHA) with the putative ferroxidase site within residues 402–417 of APP695 (Wang and Ha, 2004) (1rw6) (RMSD 0.4 Å). The Fe coordinating residues of H-ferritin, E62 and H65 (shown in red) overlap with the corresponding residues E412 and E415 that make up the putative ferroxidase site of APP (shown in green), based upon the sequence alignment in C. D & E, Kinetic values of Fe3+ formation from Fe2+ monitored by incorporation into transferrin, indicated within the graphs, were calculated for each protein (200 nM) incubated with various concentrations of Fe2+ at pH 7.2 to reflect the normal pH of brain interstitial space, where apo-transferrin is abundant (Visser et al., 2004). Cp values are in close agreement with the original reports (Osaki, 1966). Data are means± SEM, n= 3 replicates, typical of 3 experiments. See also Figure S1.