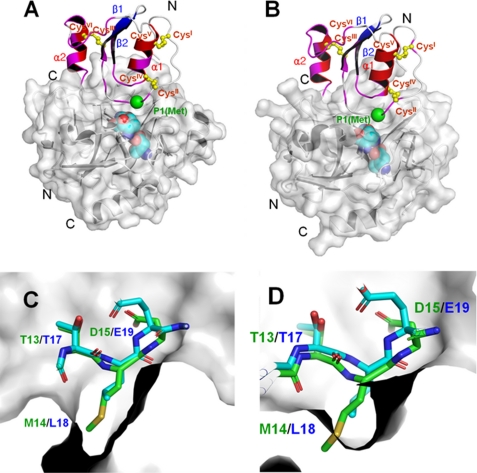
FIGURE 7.
A and B, the binding model of oryctin to α-chymotrypsin (A) and leukocyte elastase (B), predicted based on the complex structures of the OMTKY3–α-chymotrypsin complex (PDB code 1CHO) and OMTKY3-leukocyte elastase complex (PDB code 1PPF) (supplemental Fig. S5). The chemical shift-perturbed residues (>0.02 ppm) are colored in magenta. The Cα atom of the reactive site residue (P1 residue) is shown as a green sphere. The disulfide bonds are shown in yellow. The active site triads of the α-chymotrypsin and leukocyte elastase are shown as cyan spheres. A unique additional α-helix at the C terminus (α2) of oryctin also would be involved in the recognition of α-chymotrypsin and leukocyte elastase and would contribute to the binding affinity. C and D, the crystal structure of the active site loop of OMTKY3 (green) and the model of the corresponding loop of oryctin (blue) in the S1 pockets of α-chymotrypsin (C) and leukocyte elastase (D). Met14, the putative residue at the P1 position in oryctin, can fit into the S1 pocket of α-chymotrypsin, but it is too big to be accommodated by the S1 pocket of leukocyte elastase.