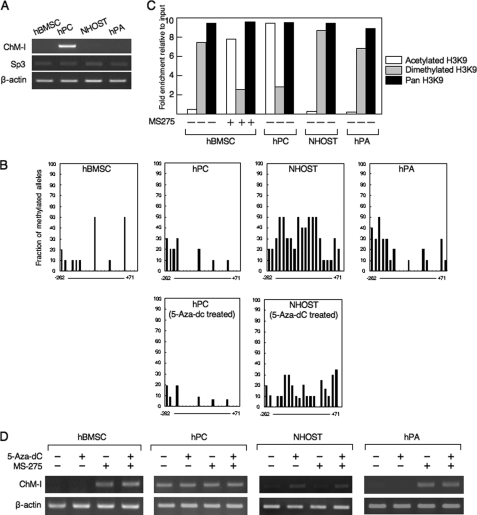
FIGURE 1.
DNA methylation and histone deacetylation down-regulate the expression of ChM-I in a cell type-specific manner. A, expression of the ChM-I and Sp3 genes in primary cultured mesenchymal cells. B, methylation status of the core-promoter region of ChM-I. The methylation of each CpG site was analyzed in 10 alleles by bisulfite genomic sequencing. The y-axis indicates the fraction of methylated alleles and the x-axis indicates the position of each CpG site relative to the transcription start site. Methylation status in hPC and NHOST treated with a demethylating reagent (5-aza-dC, 1 μm for 96 h) were also shown. C, ChIP-qPCR assay for the modification of histones in primary cultured mesenchymal cells. Open box, acetylated H3K9; closed box, dimethylated H3K9; gray box, Pan H3. Histone modification in hBMSC treated with an HDAC inhibitor (MS-275, 1 μm for 24 h) were also shown. The y-axis represents fold enrichment relative to input. D, expression of ChM-I after treatment with 5-aza-dC and/or MS-275.