Abstract
The highly complex and unique mycobacterial cell wall is critical to the survival of Mycobacteria in host cells. However, the biosynthetic pathways responsible for its synthesis are, in general, incompletely characterized. Rv3802c from Mycobacterium tuberculosis is a partially characterized phospholipase/thioesterase encoded within a genetic cluster dedicated to the synthesis of core structures of the mycobacterial cell wall, including mycolic acids and arabinogalactan. Enzymatic assays performed with purified recombinant proteins Rv3802c and its close homologs from Mycobacterium smegmatis (MSMEG_6394) and Corynebacterium glutamicum (NCgl2775) show that they all have significant lipase activities that are inhibited by tetrahydrolipstatin, an anti-obesity drug that coincidently inhibits mycobacterial cell wall biosynthesis. The crystal structure of MSMEG_6394, solved to 2.9 Å resolution, revealed an α/β hydrolase fold and a catalytic triad typically present in esterases and lipases. Furthermore, we demonstrate direct evidence of gene essentiality in M. smegmatis and show the structural consequences of loss of MSMEG_6394 function on the cellular integrity of the organism. These findings, combined with the predicted essentiality of Rv3802c in M. tuberculosis, indicate that the Rv3802c family performs a fundamental and indispensable lipase-associated function in mycobacteria.
Keywords: Bacterial Genetics, Cell Surface Enzymes, Cell Wall, Crystal Structure, Drug Action, Enzyme Inhibitors, Gene Knockout, Lipase, Mycobacterium, Tuberculosis
Introduction
The genus Mycobacterium contains a number of medically significant species, most notably the devastating human pathogen Mycobacterium tuberculosis that causes around 2 million deaths each year, the most by any single infectious agent. Despite the availability of a vaccine, the number of infected individuals worldwide continues to increase, as does the prevalence of drug-resistant forms of M. tuberculosis (1). A key virulence factor is the unique mycobacterial cell wall that consists of a core structure as follows: peptidoglycan covalently linked to arabinogalactan esterified with mycolic acids to form the mycolyl-arabinogalactan-peptidoglycan or “mAGP” complex and a series of free glycolipids, including trehalose monomycolates, trehalose dimycolates, phosphatidylinositol mannosides, and lipoarabinomannans (2), that facilitate vital interactions with host cells to initiate and maintain an infection. The essentiality of the core for mycobacterial growth and survival leads to the biosynthetic enzymes involved and being considered as ideal targets for drug development (3).
A subset of genes required for the late steps of mycolic acid and arabinogalactan biosynthesis are located in proximity to the genomes of mycobacteria and corynebacteria. These genes include a well characterized cell wall biosynthesis cluster encoding enzymes required for the activation (AccD4 and FadD32) and condensation (Pks13) (4) of mycolic acid intermediates prior to the final reduction step (5), and transfer of mature mycolic acids (6). Also within the cluster are genes required for arabinogalactan biosynthesis (atfB, glf, glfT, and Rv3806c) (7–10) which, like the mycolic acid biosynthesis genes, are essential for growth of M. tuberculosis (10).
Despite extensive functional characterization of this cluster over the last decade, the in vivo function of the product of one gene, Rv3802c, remains unknown, although mycolyltransferase (11) or Pks13-associated thioesterase (12) functions have been suggested. Although its genomic location strongly suggests a role in cell wall biosynthesis, definitive proof of such a role is lacking. The putative product of Rv3802c has a predicted signal sequence that contains a possible transmembrane domain, and it has been expressed to assess immunological responses (13) and enzymatic activities. The enzyme is one of seven cutinase-like proteins in M. tuberculosis and is retained in the cell wall, following translocation across the cell membrane (14). Previous studies have shown it to have phospholipase A and thioesterase activities (12), consistent with a role in mycolic acid biosynthesis, and significant lipase activity completely dependent on its Ser-Asp-His catalytic triad (14). A very recent study has suggested a role for Rv3802c in regulation of outer lipid composition in response to stress because the induction of the Corynebacterium glutamicum ortholog triggered an increase in mycolic acid biosynthesis as part of an outer membrane remodeling response to heat stress (15).
Recently, Rv3802c was identified as a major target of tetrahydrolipstatin (THL)6 (12). THL is a well characterized and irreversible inhibitor of serine esterases (16), originally identified for its specificity for pancreatic lipases, and thus was developed as an anti-obesity drug. THL has been reported to bind covalently to the catalytic serine residue of pancreatic lipase (17) and was found to have similar affinity for human fatty-acid synthase (18). In addition to its actions in humans, THL inhibits and disrupts cell wall formation in several mycobacterial species, with the exception of the nonpathogenic model species Mycobacterium smegmatis (19). Rv3802c was strongly inhibited by THL, whereas the nonorthologous but cutinase motif-bearing M. smegmatis lipase MSMEG_1403 was not inhibited at up to a 500:1 inhibitor to enzyme molar ratio (12).
To better understand the enzymology of Rv3802c, we expressed and purified the M. tuberculosis enzyme, its homolog in the nonpathogenic model species M. smegmatis, and the ortholog from a related species, C. glutamicum, and we assessed their inhibition by THL. We report here that the closest M. smegmatis homolog to Rv3802c, MSMEG_6394, is inhibited by THL. The crystal structure of MSMEG_6394, along with direct evidence of the essentiality of MSMEG_6394 in M. smegmatis, indicates a fundamental role of this lipase in Mycobacteria.
MATERIALS AND METHODS
Growth and Manipulation of Escherichia coli and M. smegmatis
E. coli DH5α was used for plasmid preparations during cloning experiments, although BL21-DE3 was used for protein expression. Bacteria were routinely cultured at 30, 37, or 42 °C in solid and liquid Luria Burtani (LB) medium supplemented with kanamycin (Kn, 20 μg/ml), streptomycin (Sm, 20 μg/ml), ampicillin (100 μg/ml), and sucrose (10% w/v), as appropriate. Tween 80 was added to 0.05% (v/v) to reduce clumping in mycobacterial liquid cultures. Competent M. smegmatis mc2155 cells were prepared as described previously (20) and electroporated using a Bio-Rad Gene Pulser with the following settings: 2.5 kV, 1000 ohms, 25 microfarads.
DNA Manipulations
PCRs were performed using ProofStart DNA polymerase (Qiagen) according to the manufacturer's instructions. Reactions consisted of a hot start (95 °C, 5 min) followed by 35 cycles of denaturation (95 °C, 1 min), annealing (55 °C, 1 min), and extension (72 °C, 2 min). Restriction enzymes and T4 DNA polymerase were from Roche Applied Science or New England Biolabs. Genomic DNA was prepared from mycobacteria as described previously (21). Southern blots (22) were performed using digoxygenin-labeled probes (Roche Applied Science) according to the manufacturer's instructions. M. tuberculosis H37Rv genomic DNA was obtained from Colorado State University (Fort Collins, CO).
Protein Expression and Refolding
Rv3802c was produced as described previously (14). Briefly, Rv3802c was amplified from H37Rv genomic DNA and cloned into an E. coli expression vector (pET19b; Merck). Cytoplasmic, N-terminally His-tagged recombinant protein was expressed in E. coli BL21-DE3 and accumulated in cytoplasmic inclusion bodies following induction with isopropyl 1-thio-β-d-galactopyranoside (0.5 mm). Recombinant protein was solubilized in urea and purified by immobilized metal ion affinity chromatography before refolding by dialysis into Tris (50 mm, pH 8.0).
The MSMEG_6394 gene was amplified by PCR from M. smegmatis genomic DNA using primers A (5′-GGAATTGCATATGCGCCGTCCGGACACCCC) and B (5′-CCCCAAGCTTCAACCGTGTTTCGGATGGG) and cloned into pET28b (Merck) using NdeI and HindIII (underlined). Recombinant MSMEG_6394 was expressed in B834 E. coli, and inclusion bodies were prepared as in Kjer-Nielsen et al. (23). The recombinant protein was solubilized in buffer A (20 mm Tris-HCl, 0.5 m NaCl, 8 m urea, pH 8.0) and purified by immobilized metal ion affinity chromatography. The bound protein was eluted with buffer A + 0.2 m imidazole. The eluted protein was diluted 8-fold with buffer A and refolded by dialysis against 16 liters of buffer B (10 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2 mm EDTA, 7 mm 2-mercaptoethanol) for 16 h. The refolded protein was further dialyzed against 16 liters of buffer C (10 mm Tris-HCl, pH 8.0, 20 mm NaCl, 2 mm EDTA). The refolded protein was concentrated using DEAE-cellulose (Sigma) and further purified by size-exclusion and ion-exchange chromatography.
The NCgl2775 gene was PCR-amplified from C. glutamicum ATCC 13032 genomic DNA using primers C (GGAATTGCATATGTCCGATGACTCAGATTTCATTG) and D (GCCCAAGCTTATCCGTTGTCGATGAGGTTG), digested with NdeI and HindIII (underlined), and cloned into NdeI/HindIII-digested pET28b (Merck). Primer C was designed to bind downstream of the putative signal sequence such that the first codon after the ATG start codon encoded Ser28 of NCgl2775. A sequenced clone was transformed into E. coli BL21 (DE3), and protein expression was induced at 30 °C with 1 mm isopropyl 1-thio-β-d-galactopyranoside. The recombinant NCgl2775 was found to be soluble and was purified by immobilized metal ion affinity chromatography using Talon metal affinity resin (Clontech).
Activity Assays
The esterase and lipase potentials of MSMEG_6394, Rv3802c, and NCgl2775 were determined as described previously (14, 24) with minor adaptations. The serine esterase substrate p-nitrophenyl butyrate was obtained from Sigma and was prepared in isopropyl alcohol at a range of concentrations between 200 and 1.875 mm. The substrate solutions were mixed 1:9 with a solution containing 50 mm sodium phosphate, pH 8.0, 2.3 mg/ml sodium deoxycholate, and 1 mg/ml gum arabic. To 20 μl of enzyme solution (100 μg/ml), 240 μl of the above reaction mixture was added in a 96-well microwell plate, mixed, and incubated at 37 °C for 30 min. To quantify inhibition by THL, the assay was carried out with a final p-nitrophenyl butyrate concentration of 5 mm, close to the measured Km value for each enzyme. THL was solubilized in DMSO and diluted in water to concentrations ranging between 100 μm and 400 nm. Each enzyme was preincubated with inhibitor at a 1:1 ratio for 30 min at room temperature before the addition of the substrate mixture. The accumulation of p-nitrophenol was measured spectrophotometrically at 405 nm, and concentrations were calculated by comparisons to a p-nitrophenol standard curve. All assays were performed in triplicate. Km, Vmax, Ki, and IC50 values for each enzyme were calculated using GraphPad Prism (GraphPad Software, version 4.03).
Crystallization and Data Collection
Crystals of selenomethionyl MSMEG_6394 were produced by the hanging-drop vapor diffusion method. Three μl of protein solution comprising MSMEG_6394 at 30 mg/ml, 10 mm Tris, pH 8.0, and 0.2 m NaCl were mixed with 1 μl of reservoir solution containing 2.1 m sodium formate and 0.1 m Bistris propane, pH 6.8. Crystallization trials were incubated at 21 °C. Crystals appeared after 5 days and typically grew to dimensions of 0.4 × 0.3 × 0.2 mm.
Prior to data collection, the crystals were soaked in reservoir solution with an additional 20% glycerol and flash-cooled to −173 °C in a stream of liquid nitrogen. X-ray diffraction data were collected from a single crystal mounted 300 mm from a Quantum-210 CCD detector at the BioCARS 14-BMD beamline of the Advanced Photon Source, Chicago. X-ray diffraction data were collected at wavelengths corresponding to the peak and inflection of the selenium absorption edge and at a high energy remote wavelength. The data were integrated with MOSFLM (25, 26) and scaled with SCALA (25, 27). Details of the data collection are summarized in Table 1.
TABLE 1.
Details of structural data collection and refinement
| Dataset | Inflection | Peak | Remote |
|---|---|---|---|
| Data collection | |||
| Wavelength | 0.97941 Å | 0.97930 Å | 0.96411 Å |
| Symmetry | I4122 | I4122 | I4122 |
| Cell dimensions | a = b = 130.4, c = 209.5 | a = b = 130.4, c = 209.5 | a = b =130.4, c = 209.5 |
| Resolution | 50 to 2.9 Å | 50 to 3.1 Å | 50 to 3.1 Å |
| Unique observations | 20,363 (2909) | 14,375 (2100) | 16,769 (2412) |
| Rpima | 0.025 (0.420) | 0.034 (0.305) | 0.017 (0.336) |
| Mean((I)/S.D.(I)) | 11.3 (1.8) | 9.9 (2.2) | 15.2 (1.8) |
| Completeness | 99.8% (100%) | 87.0% (88.6%) | 99.7% (100%) |
| Redundancy | 6.4 (6.3) | 4.1 (4.1) | 6.4 (6.5) |
| Refinement | |||
| Rworkb | 0.213 | ||
| Rfreeb | 0.250 | ||
| Residues | |||
| Chain A | 71–293, 298–334 | ||
| Chain B | 71–334 | ||
| Total protein atoms | 3924 | ||
| Total waters | 32 | ||
| Bond lengths | 0.016 Å | ||
| Bond angles | 1.508° | ||
| Ramachandran analysis | |||
| Favored | 91.89% | ||
| Allowed | 7.34% | ||
| Outlier | 0.77% | ||
| Average total B factor | |||
| Protein atoms | 102.5 Å2 | ||
| Average isotropic B factor | |||
| Non-protein atoms | 45.9 Å2 | ||
a Rpim = √(1/(n − 1))·Σ(|Ii − Imean|)/Σ(Ii).
b Rwork = Σhkl‖Fo| − |Fc‖/Σhkl|Fo| for all data excluding the 5% that comprised the Rfree used for cross-validation.
Structure Determination and Refinement
The structure was determined using three wavelength multiple anomalous dispersion phasing. The positions of the 18 selenium sites, corresponding to two copies of MSMEG_6394 in the asymmetric unit, were calculated using SOLVE (28), and subsequent electron density modification was performed with RESOLVE (27, 28). Into the resultant electron density map an initial peptide-backbone trace was constructed with TEXTAL (29, 30). The structure was built with iterative cycles of manual building in COOT (31) and maximum likelihood-based refinement with TLS using REFMAC (25, 32). Strict noncrystallographic restraints were maintained during refinement. The structure was validated using MOLPROBITY (33). Details of the refinement are summarized in Table 1. The coordinates and structure factors have been deposited in the Protein Data Bank under accession code 3AJA.
Construction and Analysis of a Conditional Knock-out of MSMEG_6394
MSMEG_6394 and flanking DNA were PCR-amplified from M. smegmatis mc2155 genomic DNA as a 2-kb fragment using primers E (5′-GATCAAGCTTACATGTCCGGTGAGCTGG-3′) and F (5′-GATCGGATCCGCGCACCTTGGCCCAGCG-3′), digested at the underlined restriction sites for HindIII and BamHI, and cloned into HindIII/BamHI-digested pUC18 (34). A nonpolar kanamycin resistance cassette carrying the aphA3 gene was then inserted at a unique SphI site within MSMEG_6394 after T4 polymerase treatment to form blunt ends. The 2.8-kb HindIII-BamHI fragment containing MSMEG_6394::aphA3 was then transferred to BamHI-digested pPR27, following T4 polymerase treatment of both insert and vector. To generate single crossovers, this plasmid was introduced into M. smegmatis mc2155 by electroporation and selecting kanamycin-resistant clones at 30 °C. A 10-ml LB broth containing kanamycin was inoculated with a single colony and grown for 5 days at 30 °C to saturation. Serial dilutions were plated onto LB + Kn plates at 42 °C and incubated for 4 days to select for potential single crossovers. Colonies were screened for incorporation of the plasmid into the chromosome by growing 10-ml LB + Kn cultures to saturation at 42 °C, extracting genomic DNA, digesting with XbaI/BamHI, and performing a Southern hybridization with a probe specific for MSMEG_6394. Out of nine colonies tested, one single crossover, designated Myc46, was found and subjected to further manipulation. To derive a double crossover (a conditional knock-out), a complementing plasmid, containing MSMEG_6394 carried on a 2.0-kb BamHI fragment cloned into the temperature-sensitive plasmid pCG76, was introduced into Myc46 by electroporation. Transformants were selected on LB/Kn/Sm plates at 30 °C, and a single colony was grown to saturation in LB/Kn/Sm broth at 30 °C. Serial dilutions were plated on LB/Kn/Sm plates containing sucrose and incubated at 30 °C. Potential conditional knock-out clones were grown in 10 ml of LB/Kan/Sm to saturation followed by genomic DNA extraction, digestion with XbaI/BamHI, and Southern blotting using an MSMEG_6394-specific probe. A confirmed conditional knock-out strain was designated 6394CKO. To derive growth curves, strains were cultured in 10 ml of LB/kanamycin/streptomycin at 30 °C for 3 days and then 5 ml was added to 200 ml of LB/Kan that had been pre-warmed to 30 or 42 °C. The cultures were sampled daily and serial dilutions plated onto LB/Kan at 30 °C. Following 5 days of incubation, colonies were counted to determine colony-forming units per ml.
Electron Microscopy
Bacteria were grown at 30 or 42 °C for 5 days on solid media containing appropriate antibiotics and fixed for 2 min with 2.5% glutaraldehyde in phosphate-buffered saline (PBS). Cells were scraped gently, transferred to centrifuge tubes, left in glutaraldehyde/PBS for 30 min, and centrifuged for 1 min at 5000 rpm. The solution was replaced with 2.5% glutaraldehyde and 0.05% ruthenium red in PBS and fixed overnight in the dark at 4 °C. Cells were rinsed in PBS three times for 5 min, post-fixed in 1% osmium tetroxide for 2 h, and then rinsed in water three times for 5 min. The cells were dehydrated in 10, 30, and 50% ethanol for 30 min in each concentration and then held in 70% ethanol for 4 days. Dehydration was completed in 90% ethanol for 30 min and 100% dry ethanol three times for 1 h.
Samples for TEM were then placed in propylene oxide for 1 h and then infiltrated with 25% firm grade Spurr's resin in propylene oxide for 3 days, followed by two times for 2 h in 50% Spurr's resin in propylene oxide, then 3 days in 75% Spurr's resin in propylene oxide, and 2 days in 100% Spurr's resin. Polymerization was completed at 60 °C overnight. Cells were then sectioned at 90 nm using a Reichert Ultracut S ultramicrotome and picked up onto 300 mesh copper grids. Staining was then performed with saturated uranyl acetate in 50% methanol for 10 min followed by saturated lead citrate in carbonate-free distilled water for 10 min. Sections were viewed with a Jeol 200CX TEM at 100 kV.
Samples for scanning electron microscopy were kept in 100% dry ethanol for 7 days and then rinsed in hexamethyldisilizane three times for 10 min. Drops of hexamethyldisilizane-containing cells were placed on a plastic film (unexposed, developed Ektachrome photographic emulsion) and allowed to air dry. The film was then mounted onto a double-sided carbon tape on aluminum stubs. Sputter was coated with gold using Balzers SCD 005 sputter-coating unit for 3 min at 25 mA. Cells were viewed at 10 kV and a working distance of 3 mm using a Hitachi S570 scanning electron microscope.
RESULTS
Purification of Rv3802c, MSMEG_6394, and NCgl2775
Rv3802c is very well conserved among the mycobacteria with homologs present in all genomes sequenced to date, including the minimal genome of Mycobacterium leprae (14). The M. smegmatis homolog, MSMEG_6394, shares 69% sequence identity with Rv3802c, whereas the ortholog in C. glutamicum, NCgl2775, shares 60% sequence identity with Rv3802c. The genes encoding Rv3802c, MSMEG_6394, and NCgl2775 were PCR-amplified from their respective genomes without their putative signal sequences and cloned into inducible expression vectors for production in E. coli. The construct for Rv3802c expression had been used in an earlier study and was known to yield insoluble protein that could be refolded to an active conformation (14). Expression of MSMEG_6394 in this study also gave rise to insoluble material that was successfully purified and refolded for enzymatic and structural studies. Surprisingly, NCgl2775 was found to be soluble when overexpressed in E. coli, and purified material was used directly for enzyme assays.
Enzyme Kinetics and THL Inhibition Studies
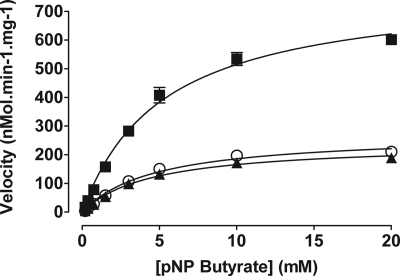
The kinetic parameters of Rv3802c and its orthologs from M. smegmatis and C. glutamicum were measured in functional assays based on the hydrolysis of p-nitrophenyl butyrate at a range of substrates. All three enzymes displayed activity under the conditions tested (Fig. 1), with MSMEG_6394 demonstrating the highest activity, with a maximum enzyme velocity of 783 nmol·min−1·mg−1 (±26.7) or more than three times that of Rv3802c. The observed specificity constants for all three enzymes were similar (Table 2).
FIGURE 1.
Kinetics of p-nitrophenyl butyrate hydrolysis. Activity of MSMEG_6394 (■), Rv3802c (▲), and NCgl2775 (○) at pH 8.0 is shown. pNP butyrate, p-nitrophenyl butyrate.
TABLE 2.
Enzyme activity of MSMEG_6394, Rv3802c, and NCgl2775 in p-nitrophenyl butyrate hydrolysis
Data shown are the mean of three experiments. Errors shown are means ± S.E.
| Vmax | kcat | Km | Specificity constant | |
|---|---|---|---|---|
| nmol·min−1·mg−1 | s−1 | mm | m−1·s−1 | |
| MSMEG_6394 | 783 ± 26.7 | 0.448 | 5.22 ± 0.45 | 86 |
| Rv3802c | 241 ± 7.8 | 0.143 | 4.52 ± 0.38 | 32 |
| NCgl2775 | 273 ± 11.8 | 0.146 | 4.71 ± 0.52 | 31 |
Activity of all three enzymes was inhibited by THL with Ki of 0.8 μm (with 95% confidence interval of 0.59–1.27) for Rv3802c (Fig. 2 and Table 3). Similar levels of inhibition were recorded whether or not samples were preincubated with inhibitors, indicating rapid and potentially irreversible inhibition. For each enzyme, 90% inhibition of activity was observed with THL concentrations less than 50 μm. This inhibition was not observed in either of the control enzymes tested, a lipase from Candida rugosa and a liver esterase from Sus domesticus (data not shown), indicating a level of specificity.
FIGURE 2.
Inhibition of enzyme activity for p-nitrophenyl butyrate by THL. Enzymes were preincubated with inhibitor for 30 min before the addition of 5 mm substrate. A, residual activity of MSMEG_6394 (○, dashed line), Rv3802c (■, solid line), and NCgl2775 (▼, solid line). B–D, curve fitting of enzyme activity for MSMEG_6394 (B), Rv3802c (C), and NCgl2775 (D) was used to calculate Ki and IC50 values.
TABLE 3.
Inhibition of p-nitrophenyl butyrate hydrolysis activity of MSMEG_6394, Rv3802c, and NCgl2775 by THL
Data shown are the mean of three experiments. Values in parentheses are 95% confidence intervals.
| IC50 | Ki | |
|---|---|---|
| μm | μm | |
| MSMEG_6394 | 5.08 (3.42–7.55) | 2.59 (1.74–3.86) |
| Rv3802c | 1.82 (1.25–2.68) | 0.87 (0.59–1.27) |
| NCgl2775 | 13.06 (6.69–25.53) | 6.34 (3.24–12.38) |
Three-dimensional Structure of MSMEG_6394
To gain functional insight into the role of Rv3802c and its orthologs, we determined the crystal structure of the M. smegmatis enzyme. Initial crystallization trials of all three proteins resulted in crystals for Rv3802c and MSMEG_6394; however, only diffraction quality crystals were obtained for MSMEG_6394. The structure of MSMEG_6394 was determined to a resolution of 2.9 Å and contained two copies of the molecule in the asymmetric unit (chains A and B). The construct used in crystallization included residues 36–337 of MSMEG_6394 as well as 20 vector-derived residues, including the His6 purification tag. Of this construct, residues 71–334 were modeled from the electron density. There was no evidence of higher order oligomerization within the crystal lattice, an observation that is consistent with the purification of MSMEG_6394 as a monomer. The protein structure confirmed that MSMEG_6394 is a member of the cutinase family of α/β hydrolases (Fig. 3A). Indeed, the closest structural homologs found with a DALI search of the Protein Data Bank were the acetyl xylan esterase (PDB code 1BS9 (35), Z-score = 19.2, r.m.s.d. = 2.6 Å over 189 Cα positions), cutinase-like protein (PDB code 2CZQ (36), Z-score = 18.8, r.m.s.d. = 2.4 Å over 185 Cα positions), lysin B (PDB code 3HC7 (37), Z-score = 18.5, r.m.s.d. = 2.9 Å over 195 Cα positions) and cutinase (PDB code 1CUS (38), Z-score = 16.1, r.m.s.d. = 2.8 Å over 173 Cα positions). All of these structural homologs shared ∼20% sequence identity with MSMEG_6394.
FIGURE 3.
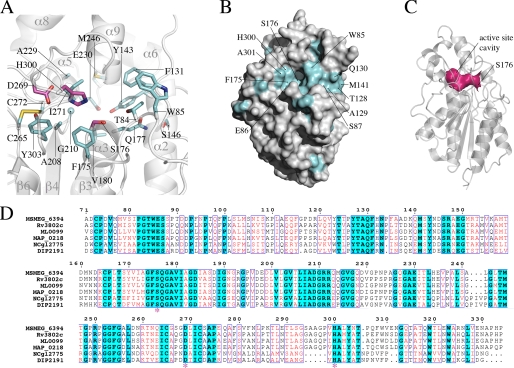
Crystal structure of MSMEG_6394. A, schematic representation of the MSMEG_6394 structure. The structure includes α/β hydrolase fold with six-stranded, parallel β-sheets (β1–β6, yellow) bounded on both sides by four α-helices (α1–α3 and α10, blue). A primarily helical “lid-like” domain (α5, α6, α8, and α9) sits atop the active site. The two disulfides in the structure are represented by green spheres. The catalytic triad residues Asp269, His300, and Ser176 are shown as magenta spheres. B, topology diagram of MSMEG_6394 colored similarly to A.
The canonical α/β hydrolase domain of MSMEG_6394 included a six-stranded, parallel β-sheet (β1–β6) bounded on both sides by four α-helices (α1–α3 and α10) (Fig. 3B). Inserted into this domain between the β5- and β6-strands and between the β6-strand and α10-helix lie a second, primarily helical domain (α5, α6, α8, and α9) atop the proposed active site. This second domain is reminiscent of the “lid” domains found in other esterase and lipase members of the α/β hydrolase superfamily and is most often associated with substrate binding specificity and interfacial activation (39).
The four cysteine residues in the protein formed two disulfide bonds, Cys73–Cys165 and Cys265–Cys272 (Fig. 3A). Both of these disulfides were structurally conserved with the acetyl xylan esterase and cutinase proteins (PDB codes 1BS9 and 1CUS), yet neither was found in lysin B (PDB code 3HC7). The first disulfide stabilizes the α/β-sandwich domain, linking the N terminus of the protein with the C-terminal region of the α2-helix. The second disulfide formed an intra-loop bridge in the β-turns spanning the β6-strand and α8-helix and is likely to be important in maintaining the conformation of the residue Asp269 from the catalytic triad.
Serine esterases typically have an active site comprising a catalytic triad of serine aspartate and histidine. The position of the catalytic triad of residues is strictly conserved between all the members of the cutinase family and coincides with the positions of Ser176, Asp269, and His300 in MSMEG_6394 (Fig. 4A). The nucleophilic Ser176 is located at the bend of the tight turn between the β3-strand and α3-helix, a feature known as the “nucleophilic elbow” that is conserved among all members of the α/β hydrolase superfamily (40). This nucleophilic elbow has the sequence Gly-Phe-Ser-Gln-Gly in MSMEG_6394, conforming to the consensus of Gly-(Phe/Tyr)-Ser-Gln-Gly with the other members of the cutinase family solved to date. Asp269 and His300 are positioned adjacent to each other, with Asp269 residing on the loop connecting the β6-strand and α7-helix and His300 within the linker between the β6-strand and α10-helix within the cutinase fold (Fig. 4A). Taken together, the structural features of MSMEG_6394 indicate that it has a catalytic mechanism akin to that of other serine esterases.
FIGURE 4.
Analysis of the active site of MSMEG_6394. A, view of the active site of MSMEG_6394 shown in the orientation depicted in Fig. 3A. The secondary structure elements are shown in schematic representation, colored gray, and labeled. Residues that are strictly conserved in the sequence alignment shown in D are represented in stick format. The catalytic triad is colored magenta, and the other conserved residues are colored cyan. B, surface representation of MSMEG_6394 orientated similarly to Fig. 3A. Residues that are strictly conserved in the sequence alignment shown in D are colored cyan and labeled. The patch of sequence conservation leading to the active site cavity may be involved in substrate recognition. C, schematic representation of MSMEG_6394 rotated 90° from the orientation shown in Fig. 3A. The solvent-accessible surface of the active site cavity is shown in magenta (analyzed with CASTp (43)). The catalytic residue Ser176 is labeled. D, sequence alignment of MSMEG_6394 homologs from related species. MSMEG_6394 is from M. smegmatis; Rv3802 is from M. tuberculosis; ML0099 is from M. leprae; MAP_0218 is from Mycobacterium paratuberculosis; NCgl2775 is from C. glutamicum, and DIP2191 is from Corynebacterium diphtheriae. The strictly conserved residues are colored cyan, and conserved residues (44) are colored red. Residues of the catalytic triad are indicated by an asterisk. Sequences were aligned with ClustalW (45) and annotated with ESPRIPT (46).
The catalytic Ser176 Oγ is positioned at the mouth of an 11-Å deep cavity that encloses the active site. This active site cavity is composed of residues from both the cutinase and lid domains of the protein and has a solvent-accessible surface area of 80 Å2 (Fig. 4, B and C). Of the residues that line the cavity, Thr84, Phe131, Tyr143, Ser176, Gln177, Val180, Ala208, Gly210, Ala229, Glu230, Met246, Ile271, and His300 are all strictly conserved across MSMEG_6394 homologs in other mycobacterial and related species (Fig. 4, A and D). The exceptions are residues 291–293 that form the C terminus of the α9-helix and line the upper part of the cavity (Fig. 4D). Similarly, there is strict conservation among mycobacterial species of the residues that form surface-exposed patches about the mouth of the active site cavity (Glu86, Phe175, His300, and Ala301) and within a patch leading to the active site (Trp85, Ser87, Thr128, Ala129, Gln130, and Met141) (Fig. 4B). The direct modeling of THL binding to MSMEG_6394 is complicated by the lack of structural homology of the lid domain between related proteins. However, the proximity of these conserved patches to the active site suggests that they, together with the residues lining the active site, may determine substrate recognition.
Conditional Disruption of MSMEG_6394 Proves Gene Essentiality in M. smegmatis
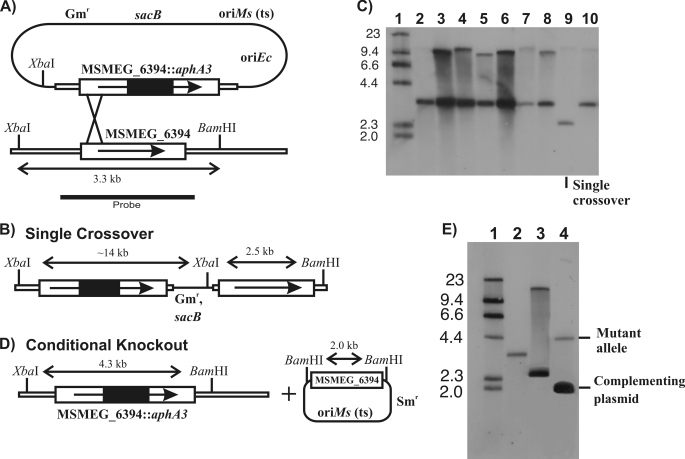
To gain insights into the function of Rv3802c, we attempted to make a mutant strain of M. smegmatis in which the MSMEG_6394 gene was disrupted with a drug resistance cassette. Despite several attempts, we were unable to generate this mutant, indicating that the enzyme might be essential to the viability of M. smegmatis and a potential drug target in mycobacteria. To investigate this further, we devised a genetic approach to assess the essentiality of MSMEG_6394 (see under “Materials and Methods”). Briefly, a homologous recombination strategy was used to disrupt MSMEG_6394 in the M. smegmatis chromosome in the presence of a rescue plasmid carrying an intact MSMEG_6394 gene (Fig. 5). This involved isolation of a single crossover strain (Fig. 5, A–C) followed by initiation of a second crossover event in the presence of a rescue plasmid encoding the MSMEG_6394 gene (Fig. 5, D and E). Allelic replacement of the chromosomal MSMEG_6394 by the disrupted copy was successfully achieved in the presence of the plasmid, giving rise to a “conditional” knock-out strain that we designated 6394CKO (Fig. 5E, lane 4).
FIGURE 5.
Construction of a conditional knock-out of MSMEG_6394. A, recombination plasmid contained a cloned copy of MSMEG_6394 interrupted by a nonpolar kanamycin resistance cassette (MSMEG_6394::aphA3), a gentamycin resistance marker (Gmr), a temperature-sensitive replication origin for M. smegmatis (oriMs (ts)), a replication origin for E. coli (oriEc), and a counterselectable marker encoding sucrose sensitivity (sacB). The construct was introduced into M. smegmatis at the permissive temperature (30 °C). Integration of the plasmid by a single crossover at the position indicated was detected by growing the cells at the nonpermissive temperature (42 °C) in the presence of kanamycin. B, genetic map of the single crossover, showing key restriction sites. C, Southern blot of XbaI/BamHI digests of genomic DNA from potential single crossover strains, probed with the fragment indicated in A. Lane 1, DNA molecular weight DNA markers of the sizes indicated in kilobases; lanes 2–10, potential single crossover strains. Lane 9 shows the single crossover strain that was selected for further manipulation. D, culturing the single crossover strain containing a complementation plasmid gave rise to a disrupted copy of MSMEG_6394 in the chromosome, producing the conditional knock-out (6394CKO). Because the disruption of MSMEG_6394 coincided with the loss of the sacB gene, the conditional knock-out strain could be selected on sucrose plates. E, Southern blot of XbaI/BamHI digests of genomic DNA showing isolation of the conditional knock-out strain. Lane 1, DNA molecular weight DNA markers of the sizes indicated in kilobases; lane 2, wild-type M. smegmatis mc2155; lane 3, single crossover strain; lane 4, conditional knock-out of MSMEG_6394 (6394CKO).
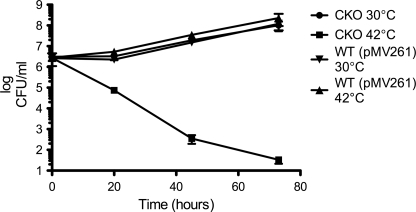
If MSMEG_6394 is essential, then 6394CKO should be fully reliant on the rescue plasmid for its survival. This plasmid has a temperature-sensitive origin of replication and can replicate at the permissive temperature (30 °C) but not at the nonpermissive temperature (42 °C) and is cured from the bacterial population when cells are grown at 42 °C (41). To confirm that 6394CKO is reliant on the rescue plasmid, the strain was cultured at 30 °C, then diluted into fresh medium at 30 and 42 °C, and sampled regularly to determine the number of viable bacteria as colony-forming units/ml. As shown in Fig. 6, 6394CKO continued to grow at 30 °C (at which the plasmid can replicate) but showed a dramatic loss of viability at 42 °C (at which the plasmid cannot replicate). By contrast, a culture of wild-type M. smegmatis mc2155 carrying the kanamycin resistance plasmid pMV261 grew well at both temperatures and at a similar rate as 6394CKO at 30 °C, showing that the loss of viability of 6394CKO at 42 °C was not just due to the temperature shift. Overall, these data confirmed that MSMEG_6394 is essential for the growth and survival of M. smegmatis.
FIGURE 6.
MSMEG_6394 is essential to the viability of M. smegmatis. The conditional knock-out strain 6394CKO was cultured at 30 °C in LB containing kanamycin and streptomycin. At saturation, 5 ml was used to inoculate 200 ml of LB/kanamycin medium that had been prewarmed at the permissive (●, 30 °C) or nonpermissive (■, 42 °C) temperature. Incubation was continued at the two temperatures, and both cultures were sampled regularly with serial dilutions plated on LB plates containing kanamycin to determine colony forming units (CFUs) per ml. A wild-type M. smegmatis mc2155 strain containing the kanamycin resistance plasmid pMV261 was included as a control (▼, 30 °C; ▲, 42°C).
Electron Microscopy Analysis of an MSMEG_6394 Conditional Knock-out
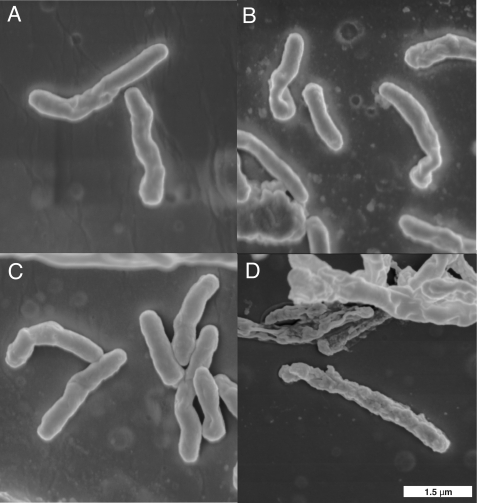
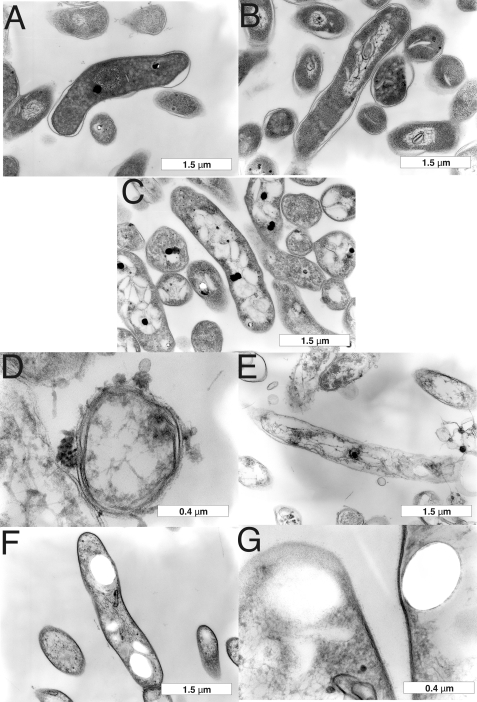
To examine the effects of the loss of MSMEG_6394 on cell structure and integrity, 6394CKO was examined by TEM and scanning electron microscopy following growth at the permissive and nonpermissive temperatures. 6394CKO was cultured for 5 days on LB agar containing kanamycin and streptomycin and then subcultured onto LB/Kan plates at 30 and 42 °C for a further 5 days to allow curing of the rescue plasmid to occur from the 42 °C samples. The cells were fixed in glutaraldehyde and then scraped from the plates and processed for microscopy (see under “Materials and Methods”). Scanning electron microscopy revealed that CKO6394 cells grown at the nonpermissive temperature (Fig. 7D) were elongated and had a rough surface relative to those grown at the permissive temperature (Fig. 7C) and wild-type controls (Fig. 7, A and B), and many appeared to have lysed. TEM was then applied to examine the cell walls and internal details (Fig. 8). TEM revealed that CKO6394 cells grown at the permissive temperature were intact with regular and typical internal compartments visible (Fig. 8C). However, CKO6394 cells grown at the nonpermissive temperature fell into one of two classes, and typical examples of each are shown in Fig. 8, D–G. Members of class 1 were elongated and showed a loss of cell wall integrity and internal structure (Fig. 8, D and E) and appeared to have lysed. Class 2 had retained an intact cell wall and internal structure (Fig. 8, F and G) but contained several electron transparent zones in their cytoplasm. In some cases, these zones were huge and dominated most of the internal space. In contrast, a wild-type strain carrying pMV261 included as a control appeared normal and intact at both temperatures (Fig. 8, A and B), showing that the cellular phenotype was not just due to the temperature shift. Combining all our findings, it is clear that MSMEG_6394 is essential for survival, playing a critical role in maintaining the cellular integrity of the bacterium. Our results have important implications regarding the role of the homologous proteins in pathogenic mycobacteria such as M. tuberculosis.
FIGURE 7.
Scanning electron microscopy of an MSMEG_6394 conditional knock-out. Bacteria were cultured on LB agar for 5 days prior to processing for scanning electron microscopy (see “Materials and Methods”). Wild-type M. smegmatis mc2155 strain containing the kanamycin resistance plasmid pMV261 was grown at 30 °C (A) or 42 °C (B). Conditional knock-out strain 6394CKO was grown at 30 °C (C) or 42 °C (D). All panels are the same magnification, and a scale bar is in the bottom right corner.
FIGURE 8.
Transmission electron microscopy of an MSMEG_6394 conditional knock-out. Bacteria were cultured on LB agar for 5 days prior to processing for TEM (see “Materials and Methods”). Wild-type M. smegmatis mc2155 strain containing the kanamycin resistance plasmid pMV261 was grown at 30 °C (A) or 42 °C (B). Conditional knock-out strain 6394CKO was grown at 30 °C (C) or 42 °C (D–G). Two classes of cells were observed for CKO6394 at 42 °C as follows: class 1 cells had lost cell wall integrity and lysed (D and E) although class 2 cells were intact but contained large electron transparent zones (F and G). Scale bars are shown at the bottom right of each panel.
DISCUSSION
The unique and highly impermeable mycobacterial cell wall is a key virulence factor that forms the interface between host and pathogen. It enables the bacterium to resist destruction by the host and also contains unusual molecules that promote uptake by host macrophages and modify host responses to create a favorable environment for bacterial survival and replication. As a result, the biosynthetic processes involved in the synthesis of the mycobacterial cell wall have been the subject of intensive research, and many of the key cell wall enzymes are now known. For example, nearly all genes within an ∼20-kb genetic locus dedicated to cell wall biosynthesis have now been characterized. In this study, we have focused on one incompletely characterized gene of this cluster, Rv3802c from M. tuberculosis.
Homology searches using M. tuberculosis Rv3802c revealed that this gene is very well conserved in Actinomycetes genomes, and we chose to focus on previously uncharacterized homologs from M. smegmatis and C. glutamicum. Previous studies using p-nitrophenyl butyrate substrates had revealed a significant lipase activity of Rv3802c (14), and THL-inhibited phospholipase A/thioesterase activities have also been reported (12). Here, we have shown that all three enzymes have significant lipase activities with similar affinity for the substrate and turnover rate. In addition, all three enzymes are inhibited by micromolar concentrations of THL. This suggests that the active sites of all three enzymes are relatively similar. Although Parker et al. (12) reported a lack of THL inhibition against an M. smegmatis enzyme, the enzyme tested (MSMEG_1403) is not the homolog of Rv3802c but rather a culture supernatant enzyme with phospholipase A activity. They suggested that a lack of MSMEG_1403 inhibition by THL is significant given that M. smegmatis growth is also not inhibited by the drug. However, our finding that MSMEG_6394 is inhibited by THL as well as being an essential enzyme in M. smegmatis would suggest that growth should be inhibited. We propose that the lack of growth inhibition in this organism is due to the inability of THL to reach its target(s) rather than a lack of activity against any particular enzyme. Because THL is active against M. tuberculosis, which shares a very similar cell wall architecture with M. smegmatis (42), lack of cell entry seems an unlikely possibility. We suggest that efflux of the drug via one of the many transporters in M. smegmatis is the most likely explanation for the lack of activity against this organism.
The annotation of Rv3802c as a putative cutinase and the observed enzyme activities of Rv3802c and its orthologs are entirely consistent with the three-dimensional structure that showed MSMEG_6394 to be a member of the cutinase family of α/β hydrolases. Interestingly, one of the closest structural matches was to mycobacteriophage lysin B (37), a novel mycolylarabinogalactan esterase that cleaves the mycobacterial cell wall, releasing free mycolic acids from the arabinogalactan layer. LysB has been proposed to facilitate mycobacterial lysis by cleaving the outer mycolate-containing layer of the cell wall from the peptidoglycan-arabinogalactan complex (37), promoting bacteriophage release from its host. The structural similarity raises the possibility of a similar function for Rv3802c, and points to a degradative role for this enzyme during mycobacterial growth. Because the deposition of new cell wall material requires cleavage and opening of the existing structure, a lipase-like Rv3802c could theoretically fulfill this role. Indeed, the unusual cell wall of members of the Actinomycetes would require the presence of a dedicated enzyme restricted to this group of bacteria. This role is consistent with the retention of Rv3802c in the cell wall, although other cutinase-like proteins are secreted into the culture filtrate of M. tuberculosis (14). This function would also explain the proposed essentiality of Rv3802c and the elongated nature of our conditional knock-out in M. smegmatis grown at the nonpermissive temperature, as described below.
Rv3802c is thought to be essential for life in M. tuberculosis because of the failure of the Rv3802c gene to accumulate transposon insertions in saturation mutagenesis experiments (10), although direct evidence of essentiality is lacking. Our inability to disrupt the homologous gene in M. smegmatis (MSMEG_6394) suggested essentiality in this species as well, and we confirmed this by creating and analyzing CKO6394, a conditional knock-out strain. CKO6394 was found to be reliant on the plasmid-encoded copy of MSMEG_6394, and curing the plasmid correlated with the appearance of surface roughness, loss of cell wall integrity, and appearance of large electron transparent zones in the cytoplasm of otherwise intact cells. Our interpretation of these observations is that the loss of the MSMEG_6394 gene results in cell death via the formation of the electron transparent zones followed by progression to the lysis stage over time. Given the suggested role of Rv3802c in mycolic acid biosynthesis (12), we tried to detect lipid bodies with a fluorescent lipid stain, but the results were inconclusive. The composition of these zones is not known, but they seem to be composed of a hard substance that resists the resin added during processing for EM. Interestingly, Kremer et al. (19) reported minor surface changes and the presence of electron-translucent bodies in THL-treated Mycobacterium kansasii, although these were confirmed to be lipid bodies and were not as well defined as the electron transparent zones described here. We also attempted to extract cell wall components following loss of the plasmid to try and detect an accumulating species, but cell death resulted in a spectrum of cell wall changes, most associated with cellular disintegration and not directly related to the loss of Rv3802c.
A recent study has suggested a role for Rv3802c and its orthologs in regulating outer membrane lipid composition under stress conditions (15). The authors found that the C. glutamicum ortholog NCgl2775 could be disrupted to give a viable mutant with no obvious alteration in cell wall composition, and we have confirmed these findings as our own mutant of C. glutamicum NCgl2775 was viable (data not shown). This study provided evidence of transcriptional induction of NCgl2775 during heat stress leading to an increase in mycolic acid biosynthesis and a decrease in phospholipid content (15). However, this finding does not account for the essential nature of MSMEG_6394 and Rv3802c in M. smegmatis and M. tuberculosis, respectively, under normal growth conditions. This discrepancy strongly suggests that a more fundamental role for Rv3802c and its orthologs remains unidentified. We propose a degradative role for the enzyme that is essential for the deposition of new cell wall material during active mycobacterial growth. The essentiality of Rv3802c and the fact its inhibitor, THL, also kills M. tuberculosis (19) make Rv3802c an attractive target to develop new antimicrobials to increase the arsenal against drug-resistant M. tuberculosis (1).
Acknowledgments
We thank Gunta Jaudzems for assistance with electron microscopy; Svetozar Kovacevic and David Lea-Smith for helpful discussions; Brigitte Gicquel for providing pPR27, and Helen Billman-Jacobe for providing pCG76.
This work was supported in part by the Australian Research Council Centre of Excellence in Structural and Functional Microbial Genomics and the National Health and Medical Research Council of Australia.
The atomic coordinates and structure factors (code 3aja) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- THL
- tetrahydrolipstatin
- Kn
- kanamycin
- Sm
- streptomycin
- PDB
- Protein Data Bank
- r.m.s.d.
- root mean square deviation
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- TEM
- transmission electron microscopy.
REFERENCES
- 1.World Health Organization (2009) in WHO Report 2009: Global Tuberculosis Control, Epidemiology, Strategy, Financing, pp. 1–314, World Health Organization, Geneva, Switzerland [Google Scholar]
- 2.Brennan P. J. (2003) Tuberculosis 83, 91–97 [DOI] [PubMed] [Google Scholar]
- 3.Brennan P. J., Crick D. C. (2007) Curr. Top. Med. Chem. 7, 475–488 [DOI] [PubMed] [Google Scholar]
- 4.Portevin D., De Sousa-D'Auria C., Houssin C., Grimaldi C., Chami M., Daffé M., Guilhot C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lea-Smith D. J., Pyke J. S., Tull D., McConville M. J., Coppel R. L., Crellin P. K. (2007) J. Biol. Chem. 282, 11000–11008 [DOI] [PubMed] [Google Scholar]
- 6.Jackson M., Raynaud C., Lanéelle M. A., Guilhot C., Laurent-Winter C., Ensergueix D., Gicquel B., Daffé M. (1999) Mol. Microbiol. 31, 1573–1587 [DOI] [PubMed] [Google Scholar]
- 7.Seidel M., Alderwick L. J., Birch H. L., Sahm H., Eggeling L., Besra G. S. (2007) J. Biol. Chem. 282, 14729–14740 [DOI] [PubMed] [Google Scholar]
- 8.Mikusová K., Yagi T., Stern R., McNeil M. R., Besra G. S., Crick D. C., Brennan P. J. (2000) J. Biol. Chem. 275, 33890–33897 [DOI] [PubMed] [Google Scholar]
- 9.Kremer L., Dover L. G., Morehouse C., Hitchin P., Everett M., Morris H. R., Dell A., Brennan P. J., McNeil M. R., Flaherty C., Duncan K., Besra G. S. (2001) J. Biol. Chem. 276, 26430–26440 [DOI] [PubMed] [Google Scholar]
- 10.Sassetti C. M., Boyd D. H., Rubin E. J. (2003) Mol. Microbiol. 48, 77–84 [DOI] [PubMed] [Google Scholar]
- 11.Takayama K., Wang C., Besra G. S. (2005) Clin. Microbiol. Rev. 18, 81–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parker S. K., Barkley R. M., Rino J. G., Vasil M. L. (2009) PLoS One 4, e4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.West N. P., Wozniak T. M., Valenzuela J., Feng C. G., Sher A., Ribeiro J. M., Britton W. J. (2008) Vaccine 26, 3853–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West N. P., Chow F. M., Randall E. J., Wu J., Chen J., Ribeiro J. M., Britton W. J. (2009) FASEB J. 23, 1694–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meniche X., Labarre C., de Sousa-d'Auria C., Huc E., Laval F., Tropis M., Bayan N., Portevin D., Guilhot C., Daffe M., Houssin C. (2009) J. Bacteriol. 191, 7323–7332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hadváry P., Lengsfeld H., Wolfer H. (1988) Biochem. J. 256, 357–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hadváry P., Sidler W., Meister W., Vetter W., Wolfer H. (1991) J. Biol. Chem. 266, 2021–2027 [PubMed] [Google Scholar]
- 18.Pemble C. W., 4th, Johnson L. C., Kridel S. J., Lowther W. T. (2007) Nat. Struct. Mol. Biol. 14, 704–709 [DOI] [PubMed] [Google Scholar]
- 19.Kremer L., de Chastellier C., Dobson G., Gibson K. J., Bifani P., Balor S., Gorvel J. P., Locht C., Minnikin D. E., Besra G. S. (2005) Mol. Microbiol. 57, 1113–1126 [DOI] [PubMed] [Google Scholar]
- 20.Jacobs W. R., Jr., Kalpana G. V., Cirillo J. D., Pascopella L., Snapper S. B., Udani R. A., Jones W., Barletta R. G., Bloom B. R. (1991) Methods Enzymol. 204, 537–555 [DOI] [PubMed] [Google Scholar]
- 21.Anderberg R. J., Strachan J. A., Cangelosi G. A. (1995) BioTechniques 18, 217–219 [PubMed] [Google Scholar]
- 22.Southern E. M. (1974) Anal. Biochem. 62, 317–318 [DOI] [PubMed] [Google Scholar]
- 23.Kjer-Nielsen L., Clements C. S., Purcell A. W., Brooks A. G., Whisstock J. C., Burrows S. R., McCluskey J., Rossjohn J. (2003) Immunity 18, 53–64 [DOI] [PubMed] [Google Scholar]
- 24.Winkler U. K., Stuckmann M. (1979) J. Bacteriol. 138, 663–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collaborative Computation Project No. 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 26.Leslie A. G. W. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, No. 26 [Google Scholar]
- 27.Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 28.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ioerger T. R., Holton T., Christopher J. A., Sacchettini J. C. (1999) Proc. Int. Conf. Intell. Syst. Mol. Biol. 130–137 [PubMed] [Google Scholar]
- 30.Ioerger T. R., Sacchettini J. C. (2003) Methods Enzymol. 374, 244–270 [DOI] [PubMed] [Google Scholar]
- 31.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 33.Richardson J. S. (2003) Methods Biochem. Anal. 44, 305–320 [PubMed] [Google Scholar]
- 34.Yanisch-Perron C., Vieira J., Messing J. (1985) Gene 33, 103–119 [DOI] [PubMed] [Google Scholar]
- 35.Ghosh D., Erman M., Sawicki M., Lala P., Weeks D. R., Li N., Pangborn W., Thiel D. J., Jörnvall H., Gutierrez R., Eyzaguirre J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 779–784 [DOI] [PubMed] [Google Scholar]
- 36.Masaki K., Kamini N. R., Ikeda H., Iefuji H. (2005) Appl. Environ. Microbiol. 71, 7548–7550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Payne K., Sun Q., Sacchettini J., Hatfull G. F. (2009) Mol. Microbiol. 73, 367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez C., De Geus P., Lauwereys M., Matthyssens G., Cambillau C. (1992) Nature 356, 615–618 [DOI] [PubMed] [Google Scholar]
- 39.Holmquist M. (2000) Curr. Protein Pept. Sci. 1, 209–235 [DOI] [PubMed] [Google Scholar]
- 40.Schrag J. D., Cygler M. (1997) Methods Enzymol. 284, 85–107 [DOI] [PubMed] [Google Scholar]
- 41.Guilhot C., Gicquel B., Martín C. (1992) FEMS Microbiol. Lett. 77, 181–186 [DOI] [PubMed] [Google Scholar]
- 42.Brennan P. J., Nikaido H. (1995) Annu. Rev. Biochem. 64, 29–63 [DOI] [PubMed] [Google Scholar]
- 43.Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., Liang J. (2006) Nucleic Acids Res. 34, W116–W118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Risler J. L., Delorme M. O., Delacroix H., Henaut A. (1988) J. Mol. Biol. 204, 1019–1029 [DOI] [PubMed] [Google Scholar]
- 45.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 46.Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]