Figure 3. Genome-wide Mapping of L1(Ta) Insertions in an Individual.
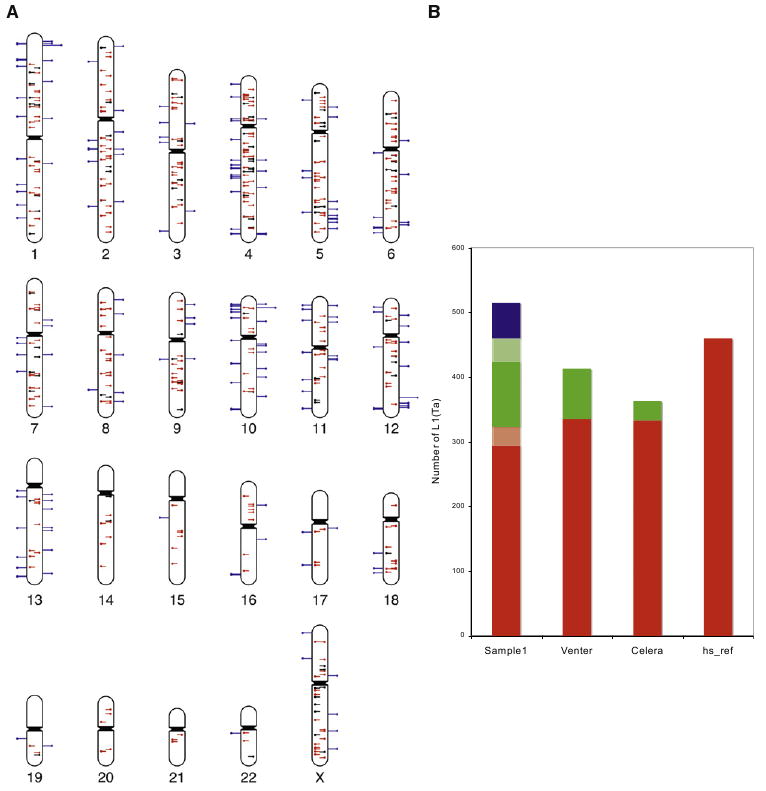
(A) Ideogram illustrates TIP-chip peaks in an individual; 514 peaks are included after imposing the cutoff (Experimental Procedures). Marks show predicted positions of L1(Ta) insertions on the plus (left side) and minus strands. Central lines similarly illustrate position and orientation of L1(Ta)s in the human reference sequence (hs_ref NCBI Build 36.1). These are color coded to indicate those identified by TIP-chip in this individual (red, n = 323) and those not seen in this sample (black). Blue lines on the outside of the chromosome correspond to nonreference insertions (n = 191). In addition to reference L1(Ta)s, 52 were considered true positives because they correspond to insertions included in dbRIP (n = 25) or were described by human sequencing projects (n = 24), as well as 3 by Beck and Moran (Beck et al., 2010). As described further in the text, additional TIP-chip peaks were verified by PCR and sequencing.
(B) TIP-chip and whole-genome sequencing in identifying L1(Ta) insertions. The y axis shows the L1(Ta) count in each sample. Sample1 was profiled by TIP-chip, whereas the other three samples are from different whole-genome sequencing approaches. Insertions present in hs_ref are displayed in red. Verified nonreference L1(Ta) insertions are shown in green. Lighter shades of red reflect reference insertions that were not retained after the imposed cutoff, while that of green reflects 3′ PCR verified insertions that might not become sequence verified. Candidate novel L1(Ta) insertions identified by TIP-chip after the cutoff and awaiting further verification, are marked in blue. The ability of TIP-chip to identify L1(Ta) insertions is comparable to whole-genome sequencing.
See also Figure S2 and Table S1.
