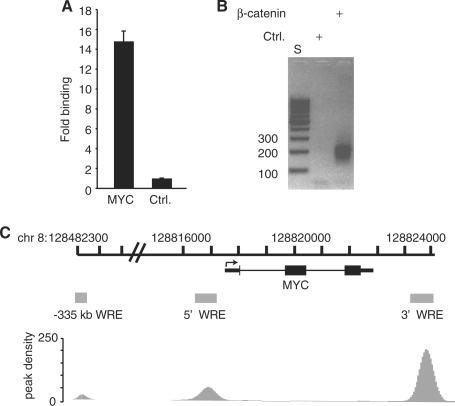
Figure 1.
Generation of a β-catenin ChIP-Seq library in HCT116 human colorectal carcinoma cells. (A) Real-time PCR analysis of DNA fragments isolated from a ChIP assay using a β-catenin specific antibody. Specific oligonucleotides were used to detect binding to a WRE downstream of the MYC transcription stop site (14,15). A region 5 kb upstream from the MYC promoter that does not associate with β-catenin/TCF4 complexes (Ctrl) served as a negative control (14). Data is presented as fold-binding relative to control and error is SEM. (B) Agarose gel analysis of size selected and PCR amplified β-catenin ChIP DNA used for Illumina high-throughput sequencing. S is a DNA standard and Ctrl is a negative control for PCR amplification (see ‘Materials and Methods’ Section). (C) Schematic of the MYC locus. Thin rectangles are untranslated regions, thick rectangles are exons, thin lines are introns and the arrow is the transcription start site. Gray shaded boxes are β-catenin enriched regions that were identified in the ChIP-Seq library which overlap 5′, 3′ and −335 kb WREs (14,22,29). Peak density plots for β-catenin enriched regions are depicted below the WREs.