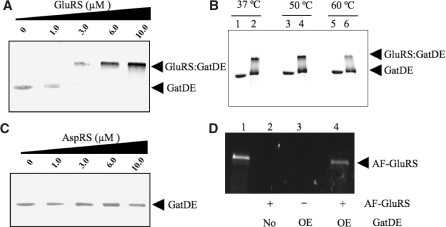
Figure 1.
Formation of the M. thermautotrophicus GluRS:GatDE binary complex. Electrophoretic mobility shift assays were performed between purified GatDE and ND-aaRS indicated as described in the ‘Materials and Methods’ section. Native PAGE (10%) separation of samples containing: (A) a stable concentration of GatDE (1 µM) and increasing concentrations of ND-GluRS (0–10 µM), (B) GatDE (3 µM) alone (lanes 1, 3 and 5) or both GatDE and GluRS (3 µM each) (lanes 2, 4 and 6) incubated at the temperatures indicated, and (C) a stable concentration of GatDE (1 µM) and increasing concentrations of ND-AspRS (0–10 µM), stained with Coomassie blue. (D) Ni-NTA pull-down with His6-tagged GatDE. Proteins were separated by SDS–PAGE and AF-GluRS was visualized by UV illumination. Escherichia coli extracts from cells either over-expressing His6-tagged GatDE (OE; lanes 3 and 4) or not (No; lane 2), were incubated with (+) or without (−) AF-GluRS (0.8 mg), and Ni-NTA agarose beads followed by three washes as described in the ‘Materials and Methods’ section. Purified AF-GluRS prior to Ni-NTA pull-down run in lane 1.