Abstract
Nuclear-encoded tRNAs are universally transcribed by RNA polymerase III (Pol-III) and contain intragenic promoters. Transcription of vertebrate tRNASec however requires extragenic promoters similar to Pol-III transcribed U6 snRNA. Here, we present a comparative analysis of tRNASec transcription in humans and the parasitic protozoa Trypanosoma brucei, two evolutionary highly diverged eukaryotes. RNAi-mediated ablation of Pol-II and Pol-III as well as oligo-dT induced transcription termination show that the human tRNASec is a Pol-III transcript. In T. brucei protein-coding genes are polycistronically transcribed by Pol-II and processed by trans-splicing and polyadenylation. tRNA genes are generally clustered in between polycistrons. However, the trypanosomal tRNASec genes are embedded within a polycistron. Their transcription is sensitive to α-amanitin and RNAi-mediated ablation of Pol-II, but not of Pol-III. Ectopic expression of the tRNASec outside but not inside a polycistron requires an added external promoter. These experiments demonstrate that trypanosomal tRNASec, in contrast to its human counterpart, is transcribed by Pol-II. Synteny analysis shows that in trypanosomatids the tRNASec gene can be found in two different polycistrons, suggesting that it has evolved twice independently. Moreover, intron-encoded tRNAs are present in a number of eukaryotic genomes indicating that Pol-II transcription of tRNAs may not be restricted to trypanosomatids.
INTRODUCTION
All eukaryotes have at least three RNA polymerases (Pol-I, Pol-II and Pol-III) that are specialized to transcribe different sets of genes (1). With few exceptions, Pol-I synthesizes the rRNAs whereas Pol-II produces mRNAs, small nuclear (sn) RNAs, small nucleolar (sno) RNAs and microRNAs. Nuclear encoded tRNAs, on the other hand, are believed to be exclusively and universally transcribed by Pol-III, which also synthesizes 5S rRNA, U6 snRNA and some other small RNAs.
Transcription of most eukaryotic tRNA genes does not require control sequences outside of the gene but depends on two intragenic promoter elements termed Box A and B. However, the tRNA that specifies selenocysteine (Sec)—the focus of our study—represents an exception. tRNASec mediates insertion of the rare amino acid Sec in response to a small number of UGA stop codons that have been recoded to Sec by the presence of a SECIS element in the 3′ UTR of a mRNA (2,3). Sec-containing proteins, termed selenoproteins, occur in all three domains of life but during evolution have been lost in some clades such as fungi and plants. Transcription of tRNASec has so far only been studied in vertebrates, and in contrast to other tRNAs it was shown to depend on three upstream regulatory regions: the TATA box motif at −30, the PSE (proximal sequence element) located around position −70 and a distal AE (activator element) located further upstream (4–7). A consensus intragenic B box can still be found whereas a functional A box is absent (Supplementary Figure S3B). The PSE and AE promoter elements are not restricted to the tRNASec gene but also occur in spliceosomal snRNA genes that are either transcribed by Pol-III (U6 snRNA) or Pol-II (U1, U2, U4 and U5 snRNAs), respectively. The PSE and the AE are known to recruit the same set of basic transcription factors for either Pol-III or Pol-II transcribed genes (8,9). Thus, it is the presence or absence of the TATA box that determines which polymerase transcribes these genes. If the TATA box is absent the gene is transcribed by Pol-II whereas if it is present, such as in the case of the tRNASec and U6 snRNA genes, it will be transcribed by Pol-III (10).
The notion that Pol-II and Pol-III independently transcribe distinct set of genes has recently been challenged. Chromatin immunoprecipitation studies have shown that in human cells Pol-II is physically associated with the upstream regions of the Pol-III transcribed U6 snRNA genes (11). Moreover, while U6 snRNAs were clearly transcribed by Pol-III their expression was inhibited by inactivation of Pol-II. More recently these results were confirmed by genome-wide study that showed that the association of Pol-II with the upstream region of Pol-III transcribed genes is not restricted to the U6 snRNA genes, but is also found for many other Pol-III transcribed genes, including tRNA genes. Furthermore, the study also showed that the same is true for many transcription factors that are usually associated with Pol-II transcription (12).
We have previously shown that the machinery and the mechanism of Sec-insertion into selenoproteins is conserved between mammals and the protozoan parasite Trypanosoma brucei (13). According to the recently revised eukaryotic phylogeny, eukaryotes are divided into six supergroups (14,15). Mammals and most of the popular model organisms such as yeast, Drosophila and nematodes belong to the supergroup of the Opisthokonta, whereas T. brucei is a representative of the supergroup of the Excavata. This suggests that the Sec-insertion machinery is conserved within eukaryotes. Thus, we wondered whether the same is true for the unusual way in which the tRNASec is transcribed. Transcription in T. brucei shows some substantial differences when compared to other eukaryotes (16–18). Trypanosomes have orthologs of all three eukaryotic polymerases (19). However, in addition to rRNAs, Pol-I also transcribes the mRNAs that code for the variant surface glycoproteins and for procyclins, the two major stage-specific surface proteins (20). Moreover, protein-coding genes are arranged in large clusters that are co-transcribed by Pol-II. Polycistronic transcription results in mRNA precursors that need to be processed into individual mature mRNAs, whereby the 5′-ends of mRNAs are formed by trans-splicing (21,22). Thus each open reading frame (ORF) of a polycistronic pre-mRNA has at least one 5′-splice acceptor site consisting of an AG dinucleotide that is preceded by a polypyrimidine tract of variable length. Both of these elements mediate the trans-splicing reaction in which a capped spliced leader (SL) sequence of 39 nt is added to the 5′-end of each mRNA [for review see (23)]. As a result, all mature mRNAs in T. brucei have an identical 39-nt SL-sequence at their 5′-end. To complete mRNA processing addition of a poly(A) tail is required. There is no consensus polyadenylation site in the 3′-untranslated region, rather polyadenylation occurs within a short region 100–400 nt upstream of the trans-splice acceptor site of the downstream gene. As in other organisms trypanosome Pol-III transcribes tRNA genes, which contain intragenic box A and B promoter elements. Unlike in other organisms, all U snRNA genes are transcribed by Pol-III, and the A and B boxes of a few tRNA genes, in addition to promoting transcription of the tRNAs, also function as upstream promoter elements for the U3 and U6 snRNA genes as well as for the 7SL RNA gene. Interestingly, in these cases the tRNAs are transcribed in the opposite directions than the three genes mentioned above (24).
In this study we performed a comparative analysis of tRNASec transcription in human cells and in T. brucei. We confirm that the human tRNASec is transcribed by Pol-III. However, transcription of the tRNASec, unlike the otherwise very similar Pol-III-mediated transcription of the human U6 snRNA genes, does not seem to require Pol-II. Surprisingly and in contrast to all other tRNAs studied so far, transcription of the tRNASec of T. brucei, is mediated by Pol-II, indicating that the mode of transcription of the tRNASec is not conserved within eukaryotes.
MATERIALS AND METHODS
Production of transgenic HeLa cells
Human tRNASec (Chr. 19, GeneID 7234) and tRNATyr (accession M55612.1) with and without T-stretch insertion (Figure 1) were overexpressed in HeLa cells using the pGEM-T Easy Vector (Promega). Each tRNA gene was expressed carrying 600 bp of its own 5′ and 300 bp of its own 3′ flanking region. HeLa cells were grown at 37°C with 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) supplemented with 10% FCS, 100 U/ml penicillin and 100 μg/ml streptomycin. Transgenic cells were obtained by seeding 1.5 × 105 HeLa cells each into six-well plates. Transfection was done the next day using 500 ng each of the tRNA expression plasmids mixed with 250 ng of the pcDNA3.1 plasmid (Invitrogen) containing an insert encoding HA-EGFP and 6 μl each of DreamFect (OZ Biosciences). One day later the efficiency of transfection (∼60%) was checked by monitoring GFP expression and total RNA was isolated as described (25). The RNAi plasmids were generated by insertion of double-stranded oligonucleotides (19 bp in length) encoding short hairpin RNAs into the pSUPERpuro vector between the BglII and HindIII sites as described previously (26). Four RNAi constructs were produced: two of them were directed against either the sequence 5′TAAAGAAGGTGAAGAACAA3′ or the sequence 5′ACATAAAGATCCCGAACAA3′ of the core subunit of human Pol-II (NM_000938.1) and two other ones targeting the core subunit of human Pol-III (NM_007055.2), either the region 5′GAGGAAATCTCTCAGGAAA 3′ or the region 5′ACGCTGAGACAGTGAGATA3′, respectively. Transgenic HeLa cells were obtained by seeding 2 × 105 cells each into six-well plates. Transfection was done the next day using 400 ng of each of the RNAi plasmids and 4 μl each of DreamFect (OZ Biosciences). One day later the cells were put under antibiotic selection by addition of 1.5 μg/ml puromycin. After 2 days of selection the antibiotic was removed and after another 24 h of incubation total RNA was isolated as described above.
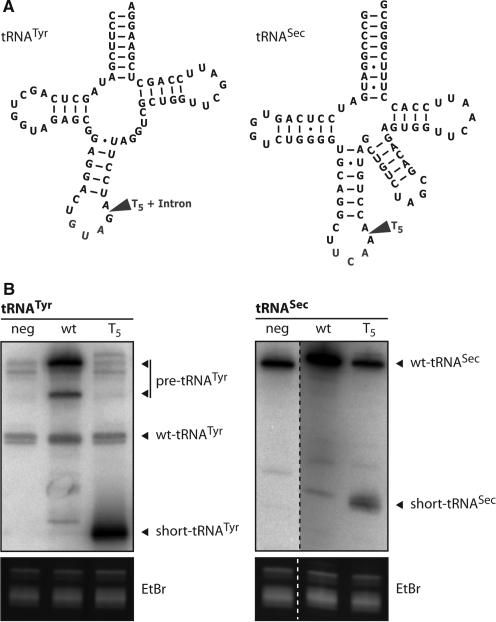
Figure 1.
An intragenic T-stretch abolishes transcription of full-length human tRNASec. (A) Predicted secondary structure of the human tRNATyr and tRNASec, respectively. Arrows indicate the positions where a synthetic Pol-III termination signal consisting of five adjacent thymidine (T5) has been inserted into the genes of the indicated tRNAs. In the case of the tRNATyr gene this position also corresponds to the position of the intron. (B) Left panel, total RNA samples isolated from control cells (neg), from transgenic cell lines overexpressing wild-type tRNATyr (wt) or the tRNATyr variant carrying the T-stretch were analyzed for the presence of tRNATyr by specific oligonucleotide hybridization on northern blots. The positions of the intron-containing tRNATyr (pre-tRNATyr), the mature wild-type tRNATyr (wt-tRNATyr) and the abortive transcript corresponding to the 5′-half of the tRNATyr (short-tRNATyr) are indicated. Right panel, same as in the left panel but analysis was done for the human tRNASec. The positions of the wild-type (wt-tRNASec) and the abortive transcript corresponding to the 5′-half of the tRNASec (short-tRNASec) are indicated. For the experiments on the left panel the cell line expressing the wild-type-tRNASec was used as a negative control (neg) whereas for the experiments on the right panel the control consisted of the cell expressing the wild-type tRNATyr. The bottom panels serve as loading controls and show the ethidium bromide stained gel segment that contains the tRNAs.
The relative RNAi-induced downregulation of Pol-II and Pol-III mRNAs when compared to the Pol-I transcribed 18S rRNA was quantified using real-time quantitative PCR (qPCR). One microgram of total RNA was reverse transcribed in 20 µl StrataScript 6.0 RT buffer containing 1 mM dNTPs, 300 ng random hexamers, 40 U RNasin (Promega), and 50 U StrataScript 6.0 reverse transcriptase (Stratagene) according to manufacturer’s protocol. Reverse transcribed material corresponding to 25 ng RNA was amplified using Brilliant® II Fast SYBR® Green QPCR Master Mix (Stratagene) and the following primers: for Pol II 5′CCAGAGCTGGAGTATCTCAGGTGTT3′ (forward) and 5′TTGCTAGCTTGCCGTCTCTACC3′ (reverse); for Pol III 5′GCTGGCTCCTGTCTACCTGTCTAAC3′ (forward) and 5′CTTGTAGCCGGCATTCAGCA3′ (reverse); 18S rRNA was detected using a commercial available TaqMan® Gene Expression Assay (Applied Biosystems, catalog no. 431-9413E) and Brilliant® II Fast QPCR Master Mix (Stratagene).
Transcription in permeabilized T. brucei cells
Transcription was analyzed using T. brucei rhodesiense Ytat 1.1 maintained at 28°C in SM medium containing 10% fetal bovine serum. We used the Ytat 1.1 strain since the analysis of transcription in permeabilized cells was originally established in this strain. Lysolecithin-permeabilized cells (27) were incubated with 32P-labeled UTP or CTP for 15 min at 28°C. Subsequently labeled RNA was isolated as described (25) and hybridized to denatured DNA spotted onto nitrocellulose membrane. Each spot contained 5 µg of DNA: the tRNASec gene and the tRNAIle gene were cloned into pTZ18U, the U6 snRNA, the tubulin and the SL genes were prepared as described (27). Membranes were hybridized overnight at 68°C in an aqueous buffer (5× SET, 10× Denhardts, 1% SDS, 10 μg/ml yeast RNA), and then washed three times for 30 min each at 68°C in 2× SSC and 0.1% SDS. Blots were exposed to a PhosphorImager screen, developed and analysed using OptiQuant software (Perkin Elmer).
Production of transgenic T. brucei cells
Pol-III activity was ablated by RNAi using a stem loop construct based on a pLew 100 (28) derivative containing the puromycin resistance gene (29). As insert we used a 480-bp fragment (nt 301–780) of the largest subunit of trypanosomal Pol-III (Tb10.70.4870). The RPB9 RNAi cell line allowing ablation of Pol-II activity was obtained from L. Vanhamme (30). Ectopic expression of tagged tRNASec and tRNAMet–i (Figure 4) was based on the same pLew100 derivative. The tagged tRNA genes were prepared by PCR mediated site directed mutagenesis. The tRNASec gene encoded on the shorter intergenic region (Figure 3) was expressed in the context of 308 nt of its own 5′ and 205 nt of its own 3′-flanking region. tRNAMet–i served as a control; it was expressed in the context of 85 bp of its own 3′-flanking region and on the 5′-side was fused to 268 bp of the 5′-flanking region of a trypanosomal tRNALeu. It has previously been shown that the tagged tRNAMet–i can efficiently be expressed in this genomic context (31). The inserts of all constructs were verified by sequencing. All transgenic cell lines are based on procyclic T. brucei 29-13 that was grown at 27°C in SDM-79 (32) supplemented with 15% FCS and the required antibiotics. Transformation, cloning and selection of transgenic cell lines were done as described (33).
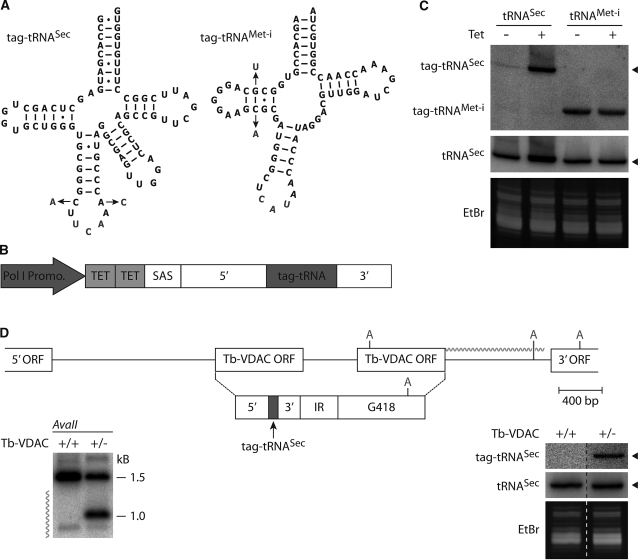
Figure 4.
Ectopic expression of the trypanosomal tRNASec gene requires an external promoter. (A) Predicted secondary structure of the tagged tRNASec and tRNAMet−i. The 2-nt changes introduced as tags are indicated. The tags allow the specific detection of the two tRNA variants by oligonucleotide hybridizations. (B) Cassette used for ectopic expression of the tRNASec (the one encoded on the shorter intergenic region) and tRNAMet−i, respectively. It contains the Pol-I procyclin promoter followed by two tetracycline operators and a splice acceptor site (SAS). The tagged tRNASec was expressed in its own genomic context, whereas the tagged tRNAMet−i was fused to the 5′-flanking region of a trypanosomal tRNALeu but retained its own 3′-flanking region (31). (C) Northern analyses of total RNA isolated from cell lines expressing the tetracycline repressor and transfected with the constructs shown in (A). tRNASec, cell line expressing the tagged tRNASec. tRNAMet−i cell line expressing the tagged tRNAMet−i. Top panel, hybridization with oligonucleotides that specifically recognize the tagged tRNASec and the tagged tRNAMet−i, respectively. Middle panel, same blot as above but reprobed with an oligonucleotide recognizing both the tagged and the endogenous tRNASec. Bottom panel, ethidium bromide stained tRNA region of the gel used for the northern analyses. (D) To scale drawing of the wild-type Tb-VDAC locus (41) and the situation after homologous recombination leading to replacement of one allele by a tRNASec/G418 resistance cassette. Relevant AvaII (A) restriction sites are indicated. The jagged line marks the probe used in the Southern analysis. Left panel: Southern analysis of genomic DNA isolated from the parental cell line (Tb-VDAC, +/+) and from a single knock out Tb-VDAC cell line (Tb-VDAC, +/−) containing the tRNASec/G418 resistance cassette. Right panel, northern blot of total RNA isolated from the same cell lines that were analyzed by the Southern blot hybridized with a probe specific for the tagged tRNASec (top panel). The same blot was reprobed with a probe recognizing both the tagged and the endogenous tRNASec (middle panel). Bottom panel, ethidium bromide stained tRNA region of the gel used for the northern analyses.
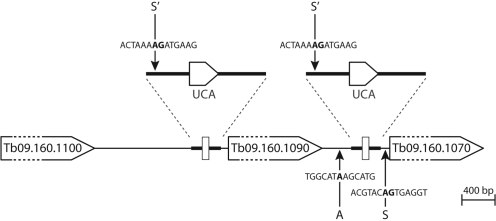
Figure 3.
Schematic illustration of the two trypanosomal tRNASec genes and their genomic context (drawn to scale). The two tRNASec genes including the flanking sequences indicated in bold are identical. Tb09.160.1090 encodes a putative serine/threonine protein kinase, the two other ORFs are annotated as hypothetical proteins of unknown function. The position and sequence of the polyadenylation site (for Tb09.160.1090) and the splice acceptor site (for Tb09.160.1070) as determined by 3′ and 5′ RACE are indicated by A and S, respectively. The functional splice acceptor site detected by deep sequencing of a poly(A) enriched SL-containing cDNA library of procyclic and bloodstream T. brucei is indicated by S′.
RT–PCR analysis in T. brucei
RT analysis to determine the splice acceptor and polyadenlyation sites was performed using the ImProm-II system (Promega) following the manufacturers procedure. Primers for the 3′-RACE of the Tb09.160.1090 encoding cDNA were: 5′TTGAATTCGCATTGAGCACCTGCTTTTTTTTTTTTTTTTTTVNN3′ (1. PCR, reverse), 5′TTGAATTCGCATTGAGCACCTGC3′ (2. PCR, reverse), 5′AAGAGCTAGAAGCACGCGG3′ (1. PCR, forward), 5′ACCAATTTCTTCATCCATTCACA3′ (2. PCR, forward). Primers for the 5′-RACE of the Tb09.160.1070 encoding cDNA: 5′TGAAACTCCATGTATTGCCGC3′ (1. PCR, reverse), 5′CTGAGGGACGACAGAGCG3′ (2. PCR, reverse), 5′CGCTATTATTAGAACAGTTTCTGTAC3′ (1. And 2. PCR forward).
Northern analysis
Denaturing polyacrylamide gels (Figures 1, 2, 4 and 6) were processed for northern blot analysis as described (34,35). The indicated 32P-5′-end labeled oligonucleotides were used as probes (T. brucei: Tb, H. sapiens: Hs): 5′ ACCAGCTGAGCTCATCGTGGC3′(Tb-tRNASec), 5′TACGGGGTTGAATCCCGCA3′ (Tb-tagged tRNASec) 5′TGCTCCCGGCGGGTTCGAA3′ (Tb-tRNAIle), 5′CGCTCTTCCCCTGAGCCA3′ (Tb-tagged tRNAMet−i), 5′CAGATTCCCGCAGTATGCGG3′ (Tb-SL RNA), 5′ACCACTGAGGATCATCCGGGC3′ (Hs-tRNASec), 5′GCTCTACCAGCTGAGCTATCG3′ (Hs-tRNATyr), 5′GTATATGTGCTGCCGAAGCGAG3′ (Hs-U6 snRNA), 5′GTGCACCGTTCCTGGAGGTACT3′ (Hs-U2 snRNA). Signals were quantitated as described above.
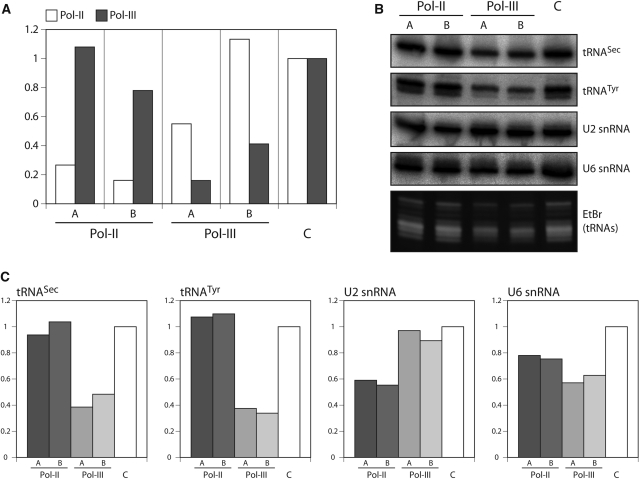
Figure 2.
RNAi of Pol-III inhibits expression of human tRNASec. (A) Real-time qPCR analysis of the mRNA levels of the core subunit of Pol-II (white bars) or Pol-III (grey bars) from HeLa cells transfected with either the empty plasmid (C) or with plasmids expressing shRNAs targeting the mRNAs of the core subunit of Pol-II or Pol-III, respectively. For both RNA polymerases, cells were transfected with two independent shRNA expressing plasmids that target distinct regions (A and B) of the corresponding mRNAs. All mRNAs levels have been normalized to the level of the 18S rRNA. The levels of mRNAs encoding the core subunits of Pol-II and Pol-III in the cell line transfected with the empty plasmid (C) was set to 1. (B) Northern analysis of 4 µg of total RNA isolated from control cells and from cells undergoing RNAi. Four days after transfection total RNA was isolated and analyzed for the presence of the indicated RNAs by specific oligonucleotide hybridization. The bottom panels serve as loading controls and show the ethidium bromide stained gel segment that contains the tRNAs. (C) Quantification of the results shown in (B). The signal in the control cells (C) that do not express shRNAs was set to 1.
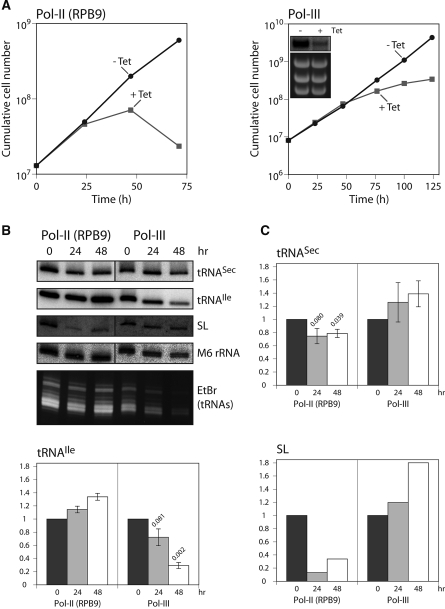
Figure 6.
Effect of RNAi-mediated ablation of Pol-II and Pol-III activities on steady state levels of different RNAs. (A) Growth of uninduced and induced procyclic T. brucei RNAi cell lines downregulated for RPB9 (30) or the largest subunit of Pol-III. Inset: downregulation of the mRNA encoding the largest subunit of Pol-III was verified by northern blot analysis 24 h after induction of RNAi. (B) Northern analyses of total RNA isolated from uninduced (0) and induced (24, 48 h) RPB9 and Pol-III RNAi cell lines. RNA was resolved in 8 M urea on a 10% polyacrylamide gel and hybridized with oligonucleotides specifically detecting the transcripts indicated on the right. The tRNA region of the corresponding ethidium bromide stained gel is shown at the bottom. (C) Quantitation of the northern blots shown in (B). Signals were normalized to the cytosolic M6 rRNAs not affected by ablation of either RPB9 or Pol-III. For tRNASec and tRNAIle the means of three experiments are shown. The signal in the uninduced cells was set to 1. Standard errors and the relevant P-values (Student’s t-test, one-tailed, paired) are indicated.
RESULTS
An intragenic T-stretch abolishes transcription of full-length human tRNASec
Transcription of vertebrate tRNASec and the multiple U6 snRNA genes (36) is very similar: both require a TATA box, the PSE and the AE and share several transcription factors. However, in the case of the tRNASec surprisingly little attention has been paid to the question whether it is transcribed by Pol-III. Early experiments showed that transcription of the tRNASec is resistant to 1 µg/ml of α-amanitin suggesting that it is transcribed by Pol-III (4,6). However, it was not shown that under these conditions Pol-II-dependent transcription was inhibited. Moreover, it is known that short T-stretches act as termination signals for Pol-III. Consequently, tRNA genes are generally flanked at their 3′-end by short T-stretches. However, in case of the human tRNASec the first T-stretch (T4) occurs only 45-nt downstream of the 3′-end of the gene. Finally, recent studies with human U6 snRNA have shown that even though its transcription is mediated by Pol-III, it also depends on active Pol-II (11). In summary, these observations underscore the complexity of tRNASec and U6 snRNA transcription modes and prompted us to reinvestigate whether human tRNASec is indeed transcribed by Pol-III. Practically this was achieved by using novel methods that for the most part were not available at the time the pioneering studies mentioned above were performed (4,6).
In order to identify the polymerase responsible for transcription of human tRNASec, we designed a tRNASec gene variant containing a Pol-III termination signal consisting of five thymidine residues at position 37 in the region of the tRNA that encodes the anticodon loop (Figure 1A). If Pol-III transcribes the variant tRNASec, transcription should prematurely terminate within the T-stretch, whereas if the tRNA is transcribed by Pol-II the full-length tRNA carrying the T-stretch insertion should be obtained. To show that the assay works as intended, we also designed a human tRNATyr gene variant containing five thymidines at the same relative position as in the variant tRNASec gene (Figure 1A). In the tRNATyr gene, this position coincides with the 5′-end of its intron. Constructs containing the corresponding wild-type tRNA genes and their variants containing the synthetic T-stretch insertions were transfected into HeLa cells. Twenty-four hours later expression of the transgenes was analyzed by northern blotting. Overexpression of wild-type tRNATyr gene resulted in the accumulation of large amounts of intron-containing precursor tRNATyr and, to a lesser extent, of the spliced form (Figure 1B). The accumulation of precursor rather than mature tRNATyr is most likely caused by overloading of the tRNA splicing machinery. Overexpressing the tRNATyr variant, on the other hand, resulted in the accumulation of large amounts of a shorter fragment, whose size is consistent with a molecule representing the 5′-half of the tRNATyr, extended by an unknown number of T residues derived from termination within the T-stretch insertion (Figure 1B), as expected for a Pol-III transcript, The analogous experiments were also done for the human tRNASec. The extent of overexpression was less than what was observed for the tRNATyr. However, also in this case the T-stretch containing tRNASec variant gene led to the accumulation of a short fragment, whose length also corresponds to the expected length of the 5′-half of the tRNASec carrying the T-stretch nucleotides. Furthermore, no band corresponding to the full length tRNASec containing the added 5-nt long T-stretch was detected. These results show that human tRNASec responds to a typical Pol-III termination signal and therefore suggest that it is transcribed by Pol-III.
RNAi of Pol-III inhibits expression of human tRNASec
It recently became clear that both Pol-III and Pol-II are required for efficient expression of the human U6 snRNA genes (11). In order to analyze whether this might also be the case for the human tRNASec gene, we performed RNAi experiments. HeLa cells were transfected with constructs allowing overexpression of short hairpin (sh) RNAs designed to induce RNAi-mediated ablation of the Pol-II or Pol-III core subunits, respectively. For each of the polymerases, the knockdown was performed independently with two shRNA expressing plasmids targeting different regions of the corresponding mRNAs. Following transfection, the cells were incubated for two days in the presence of puromycin in order to enrich for cells expressing the transgenic shRNAs. Figure 2A shows that, as expected, the Pol-II or Pol-III mRNA levels were specifically downregulated in the corresponding RNAi cell lines but not in cells transfected with a control plasmid. Pol-II and Pol-III are essential, and RNAi directed against the corresponding mRNAs is therefore expected to be lethal. Thus, RNA was isolated and analyzed 4 days after transfection at the time point when the cells were still completely viable. Figure 2 shows that ablation of Pol-II did neither affect the level of tRNASec nor the one of tRNATyr. However, consistent with the notion that both tRNAs are transcribed by Pol-III their levels dropped to ∼40% in cell lines downregulated for Pol-III. As control for a Pol-II transcript we analyzed the levels of the U2 snRNA which were reduced to ∼55% in the Pol-II RNAi cell lines but unaffected by ablation of Pol-III. Finally, we also analyzed the Pol-III transcribed U6 snRNA. Interestingly, its expression was reduced to ∼45% in Pol-III RNAi cell lines and also by 25% in the Pol-II RNAi cells. This latter result supports the recent suggestion that while human U6 snRNA is transcribed by Pol-III, its expression also requires active Pol-II (11). Moreover, the results confirm that human tRNASec is transcribed by Pol-III and suggest that, while there are many shared features between the transcription mode of the tRNASec and the U6 snRNA, the requirement for an active Pol-II is not one of them.
Unique tRNASec gene loci in T. brucei
Previous work has shown that the machinery and the mechanism of Sec-insertion into selenoproteins is conserved between mammals and T. brucei (13). This raises the question whether this conservation also extends to the transcription of tRNASec genes. The large majority of trypanosomal tRNA genes occurs in clusters of two to five or more genes separated by short intergenic regions. Within clusters, tRNA genes can be arranged in head-to-head, tail-to-tail or head-to-tail orientations (34,37). Trypanosomal tRNA gene clusters are confined to genomic regions of at least 5 kb in length which are devoid of protein-coding genes (this is also true for the few tRNA genes that do not occur in clusters). Some, but not all, tRNA gene-containing loci coincide with strand switch regions where the transcription of two polycistronic transcription units either converges or diverges (38). The T. brucei genome encodes two identical tRNASec genes, which unlike all other tRNAs are found in the intergenic regions (1640 and 833 bp in length, respectively) between three adjacent protein-coding genes of a polycistronic transcription unit (Figure 3).
Sequence analysis indicates that the trypanosomal tRNASec genes appear to lack consensus sequences for the internal A and B boxes, as well as any upstream Pol-III promoter elements. The peculiar genomic arrangement, together with the apparent absence of functional promoter elements, suggest that the trypanosomal tRNASec genes might be transcribed as part of a polycistronic RNA in conjunction with the protein-coding genes.
This is further supported by the fact that no histone variants, that mark the boundaries of polycistronic transcription units in T. brucei, are found in chromatin associated with the tRNASec genes (39).
To analyze the situation in more detail for the tRNASec gene situated within the shorter intergenic region we determined the polyadenylation site of the upstream mRNA and the splice acceptor site of the downstream mRNA, respectively (Figure 3). Moreover, using RT–PCR we showed that the whole intergenic region together with the flanking ORFs is transcribed as one RNA molecule (Supplementary Figure S1). Interestingly, deep sequencing of poly(A)-enriched SL-containing cDNA libraries of procyclic and bloodstream T. brucei revealed a small number of molecules consisting of the SL sequence attached to the 5′-flanking region of the two tRNASec genes (data not shown). This suggests that the transcripts that contain the tRNASec are processed from a polycistronic precursor like mRNAs by trans-splicing and polyadenylation. The resulting trans-spliced RNA fragments could then be further processed like other tRNA precursors by RNAse P and a combination of endo- and exonucleases, respectively. This situation would be reminiscent of the Pol-II-transcribed snoRNAs of other organisms that are frequently encoded in introns and nucleolytically processed after splicing (40).
Ectopic expression of trypanosomal tRNASec requires an external promoter
If the trypanosomal tRNASec genes lack a Pol-III promoter and instead are transcribed by Pol-II, they should be transcriptionally silent when integrated outside of a polycistron. To test this prediction we integrated a tagged version of one of the two tRNASec genes (Figure 4A) including its flanking regions and an upstream tetracycline (tet)-inducible Pol-I promoter into a non-transcribed intergenic rDNA region (Figure 4B). Figure 4C shows that in this genomic context expression of the tagged tRNASec depended on the presence of tet, indicating that this gene does not contain an internal promoter. A weak signal is detected in uninduced cells, which could be due to leaky Pol-I transcription in the absence of tet. It should be mentioned here that the intensity of this signal was always very weak and for unknown reasons somewhat variable. Supplementary Figure S4 shows an example of an experiment where tRNASec expression was strictly tet dependent. Moreover, the experiments in Figure 4C show that the Pol-I-transcribed tRNASec is correctly processed since it co-migrates with the endogenous wild-type tRNASec (Figure 4C, middle panel). As control we tested expression of a tagged initiator tRNAMet (31) in the context of the same construct. In this case, as expected for Pol-III-directed transcription, expression was constitutive and therefore independent of an active Pol-I promoter (Figure 4C).
Integration of the tRNASec gene into a polycistron is sufficient for its expression
The experiments described in Figure 4C raise the question whether the tRNASec gene would be expressed when placed in a different polycistron than the one it naturally resides in. In order to test this hypothesis we transplanted the same tagged tRNASec gene cassette that was used for the experiment in Figure 4C into the polycistron that encodes the Tb-VDAC (voltage-dependent anion channel) genes. This polycistron was chosen since previous studies have shown that the VDAC genes are not essential for normal growth of T. brucei in SDM-79 medium (41). The Southern analysis in the left panel of Figure 4D shows that, as expected, the cassette consisting of the tRNASec gene followed by the G418 resistance gene replaced one allele of the Tb-VDAC locus. Subsequent northern analysis using a specific oligonucleotide probe showed that the tagged tRNASec is expressed and correctly processed under these conditions (Figure 4D, right panel). Thus, whereas ectopic expression of the tRNASec gene outside of a polycistron requires an external promoter (Figure 4C) placing it into a polycistron allows it to be expressed. This observation strongly suggests that tRNASec is transcribed by co-expression with Pol-II transcribed protein-coding genes.
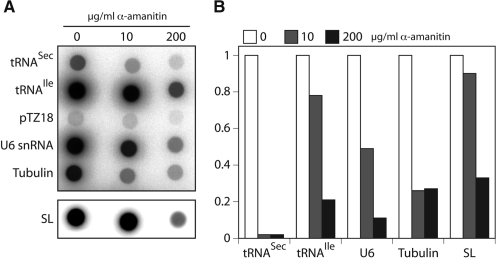
α-Amanitin inhibits transcription of trypanosomal tRNASec
To determine which RNA polymerase is responsible for transcription of the trypanosomal tRNASec genes we performed nascent transcript labeling assays in lysolecithin-permeabilized cells (27). RNA from untreated and α-amanitin-treated cells was synthesized in the presence of radioactive UTP or CTP and hybridized to dot blots containing Pol-II and Pol-III transcribed genes. Figure 5 shows that maximum inhibition of transcription of the tRNASec genes as well as of the Pol-II-transcribed tubulin genes is reached at 10 µg/ml α-amanitin. In contrast, expression of the Pol-III-transcribed tRNAIle and U6 snRNA is to a large extent resistant to 10 µg/ml α-amanitin but dramatically reduced at 200 µg/ml. Moreover, as shown previously, transcription of the SL RNA gene is less sensitive to α-amanitin than other Pol-II-transcribed genes (27). Sensitivity to low concentration of α-amanitin is a hallmark of Pol-II-mediated transcription and suggests that the trypanosomal tRNASec is transcribed by Pol-II.
Figure 5.
Transcription of the tRNASec gene is α-amanitin-sensitive. (A) UTP-labeled RNA was synthesized in permeabilized trypanosomes in the absence and presence of 10 and 200 µg/ml of α-amanitin and hybridized to nitrocellulose filters containing immobilized cloned DNA of the indicated genes (pTZ18: vector control; SL: spliced leader). (B) PhosphorImager quantitation of the results as shown in panel (A). The samples without α-amanitin were set to 1. Values represent means of two experiments. The variation between the values of two experiments was <15%.
RNAi of Pol-II but not of Pol-III affects transcription of trypanosomal tRNASec
The question of which RNA polymerase transcribes trypanosomal tRNASec was also addressed in a manner analogous to the experiment with human cells shown in Figure 2. Thus, we used RNAi cell lines allowing inducible downregulation of subunit RPB9 of Pol-II (30) or of the largest subunit of Pol-III, respectively. Induction of the RPB9-RNAi cell line with tet leads to a growth arrest after ∼24 h and subsequent cell death. Ablation of the largest subunit of Pol-III causes a slow growth phenotype after ∼48 h (Figure 6A). Changes of the steady-state levels of tRNASec, Pol-III-transcribed tRNAIle as well as Pol-II-transcribed SL RNA were analyzed in uninduced and induced RNAi cell lines by northern blotting (Figure 6B). The steady-state levels of each of the tested RNAs were quantified after normalization to a Pol-I-transcribed small rRNA (M6 rRNA), which was not affected in the two RNAi cell lines. Figure 6C shows that the level of the tRNASec was reduced by ∼25% in the RPB9-RNAi cell line but actually increased in the Pol-III RNAi cell line. The SL RNA showed a similar but more pronounced behavior: its level was reduced in the Pol-II RNAi cell line to ∼30% and increased in the Pol-III RNAi cell line. The converse result was obtained for the tRNAIle: its level increased by ∼30% in RPB9-RNAi cells and was reduced to ∼30% after downregulation of Pol-III. The ethidium bromide stained gel (Figure 6B, bottom panel) shows that the bulk of tRNAs was downregulated by Pol-III knockdown. Unexpectedly, all tested RNA species showed an increase in their steady levels in the RNAi cell line ablated for the RNA polymerase that should not affect their transcription. This is potentially highly interesting since it may indicate an as yet unknown cross talk between the two RNA polymerases. It could, for example, be that both RNA polymerases compete for a common rate-limiting transcription factor. However, it is important to emphasize that ablation of either RNA polymerase is expected to have dramatic consequences on the physiology of the cells. The interpretation of these unexpected results is therefore very difficult and requires more elaborate investigations that are beyond the scope of this study.
However, in summary, the RNAi results are consistent with the experiments in permeabilized cells (Figure 5) and show that trypanosomal tRNASec is transcribed by Pol-II.
DISCUSSION
Eukaryotes are divided into six supergroups: humans belong to the Opisthokonta and T. brucei to the Excavata. Only a few species outside the opisthokont clade are known to have a tRNASec. These include Chlamydomonas reinhardtii, Dictyostelium discoideum, Tetrahymena and members of the apicomplexans such as Plasmodium falciparum and Toxoplasma gondii. These species represent three supergroups: the Archeaplastida (C. reinhardtii), the Amoebozoa (D. discoideum) and the Chromalveolata (Tetrahymena and apicomplexans). In none of these species transcription of the tRNASec gene has been analyzed. However, sequence comparison shows that the putative tRNASec B box region (37,42) is conserved in most species, as would be expected if the gene is transcribed by Pol-III. The only exceptions are T. brucei, L. major and D. discoideum (Supplementary Figure S2). The fact that the tRNASec gene of D. discoideum, similarly to the ones of trypanosomatids, appears to have a deficient B box indicates that Pol-II transcription of tRNASec may occur in the Archeaplastida. However, without experimental evidence it remains possible that the B box of the D. disciodeum tRNASec is functional even though it deviates from B box consensus sequence. In summary, the sequence analysis shown in Supplementary Figure S2 suggests that Pol-III-mediated transcription of tRNASec represents the ancestral situation and that Pol-II-mediated transcription of tRNASec, as observed in trypanosomatids, is a derived trait. It should be emphasized in that context that as far as we can tell all other trypanosomal tRNAs are transcribed by Pol-III.
Did this shift to Pol-II-mediated transcription of tRNASec occur by random genetic drift in the ancestor of trypanosomatids or does it reflect an adaptation that has functional importance? Selenoprotein synthesis requires at least four dedicated factors: phosphoseryl-tRNASec kinase, phosphoseryl-tRNA:selenocysteinyl-tRNA synthase, tRNASec-specific elongation factor and selenophosphate synthetase (13,43). The synteny of the genes encoding these four proteins is conserved in the trypanosomatid species T. brucei, T. cruzi and Leishmania major (44). However, this is not the case for the tRNASec genes: they are found within a polycistron in all three species, but the flanking genes in L. major are different from those in T. brucei and T. cruzi (Supplementary Figure S3A). This genetic arrangement shows that integration of the tRNASec gene into a polycistron is a relatively recent event that evolved twice independently in the trypanosomatid lineage and suggests that is has been positively selected for.
Pol-III-mediated transcription of eukaryotic tRNASec genes shows some peculiarities. Unlike other tRNA genes it requires upstream promoter elements (Supplementary Figure S3B) (5,45). These elements are very similar to the ones required for transcription of the U6 snRNA genes (9). It is interesting to note that transcription of the trypanosomal U6 snRNA gene, while still dependent on Pol-III, is different than in other eukaryotes since it requires an upstream inverted tRNA gene as a promoter element (Supplementary Figure S3C) (24). Why trypanosomal U6 snRNA is transcribed in a different way than in other systems is not known, but it should be considered that trypanosomatids appear to lack many transcription factors that are conserved in other eukaryotes (46). Thus, it is possible that the Pol-II-mediated transcription of the trypanosomal tRNASec is connected to the fact that the standard eukaryotic U6 snRNA transcription mode is not operational in T. brucei. Most of the T. brucei genome codes for large Pol-II-transcribed polycistrons which may have predisposed the tRNASec gene to be transcribed by Pol-II. All that needs to be postulated for such a scenario is the integration of the tRNASec gene into a polycistron.
tRNASec is unusual in many respects, so we wondered if there was any evidence for Pol-II-mediated transcription of conventional tRNAs. Analyzing the genomic organization of tRNA genes in Caenorhabdititis elegans (http://www.wormbase.org, release WS201) and Drosophila melanogaster (http://flybase.org/) we found in both organisms a few cases where tRNAs are encoded within introns of Pol-II-transcribed genes. Interestingly, in C. elegans one of the tRNA genes that is found within an intron is tRNASec (http://www.wormbase.org/db/get?name=WBGene00023106;class=Gene), although unlike in trypanosomatids it is not present in a polycistron. Intron-encoded tRNA genes are embedded in Pol-II transcription units and therefore must be transcribed by Pol-II, although it is possible that they still contain internal Pol-III promoters and may be independently transcribed by Pol-III. That Pol-II-mediated transcription of tRNAs is in principle possible has recently been shown in mouse where insertion of a circularily permuted tRNA gene into a protein-coding gene yielded a functional tRNA, a large fraction of which was transcribed by Pol-II (47). Thus these findings indicate that Pol-II transcription of tRNA genes may not be restricted to trypanosomatids but be more widespread in eukaryotes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Foundation (grant number 121937 to A.S.), (grant number 112092 to I.R.) and (grant number 113878 to O.M.); National Institutes of Health (grant number AI028798 to E.U.). Funding for open access charges: University of Berne.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank S. Devaux and L. Vanhamme for providing them with the RPB9 RNAi knockdown cell line.
REFERENCES
- 1.Archambault J, Friesen JD. Genetics of eukaryotic RNA polymerasese I, II and III. Microbiol. Rev. 1993;57:703–724. doi: 10.1128/mr.57.3.703-724.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Commans S, Böck A. Selenocysteine inserting tRNAs: an overview. FEMS Microbiol. Rev. 1999;23:335–351. doi: 10.1111/j.1574-6976.1999.tb00403.x. [DOI] [PubMed] [Google Scholar]
- 3.Hatfield DL, Carlson BA, Xu XM, Mix H, Gladyshev VN. Selenocysteine incorporation machinery and the role of selenoproteins in development and health. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:97–142. doi: 10.1016/S0079-6603(06)81003-2. [DOI] [PubMed] [Google Scholar]
- 4.Lee BJ, Kang SG, Hatfield D. Transcription of Xenopus selenocysteine tRNA Ser (formerly designated opal suppressor phosphoserine tRNA) gene is directed by multiple 5′-extragenic regulatory elements. J. Biol. Chem. 1989;264:9696–9702. [PubMed] [Google Scholar]
- 5.Myslinski E, Krol A, Carbon P. Optimal tRNA(Ser)Sec gene activity requires an upstream SPH motif. Nucleic Acids Res. 1992;20:203–209. doi: 10.1093/nar/20.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carbon P, Krol A. Transcription of the Xenopus laevis selenocysteine tRNA(Ser)Sec gene: a system that combines an internal B box and upstream elements also found in U6 snRNA genes. EMBO J. 1991;10:599–606. doi: 10.1002/j.1460-2075.1991.tb07987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuster C, Myslinski E, Krol A, Carbon P. Staf, a novel zinc finger protein that activates the RNA polymerase III promoter of the selenocysteine tRNA gene. EMBO J. 1995;14:3777–3787. doi: 10.1002/j.1460-2075.1995.tb00047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jawdekar GW, Henry RW. Transcriptional regulation of human small nuclear RNA genes. Biochim. Biophys. Acta. 2008;1779:295–305. doi: 10.1016/j.bbagrm.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hernandez N. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J. Biol. Chem. 2001;276:26733–26736. doi: 10.1074/jbc.R100032200. [DOI] [PubMed] [Google Scholar]
- 10.Lobo SM, Hernandez N. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell. 1989;58:55–67. doi: 10.1016/0092-8674(89)90402-9. [DOI] [PubMed] [Google Scholar]
- 11.Listerman I, Bledau AS, Grishina I, Neugebauer KM. Extragenic accumulation of RNA polymerase II enhances transcription by RNA polymerase III. PLoS Genet. 2007;3:e212. doi: 10.1371/journal.pgen.0030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Struhl K, Snyder M. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc. Natl Acad. Sci. USA. 2010;107:3639–3644. doi: 10.1073/pnas.0911315106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aeby E, Palioura S, Pusnik M, Marazzi J, Lieberman A, Ullu E, Söll D, Schneider A. The canonical pathway for selenocysteine insertion is dispensable in Trypanosomes. Proc. Natl Acad. Sci. USA. 2009;106:5088–5092. doi: 10.1073/pnas.0901575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 15.Simpson AG, Roger AJ. The real ‘kingdoms' of eukaryotes. Curr. Biol. 2004;14:R693–R696. doi: 10.1016/j.cub.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 16.Campbell DA, Thomas S, Sturm NR. Transcription in kinetoplastid protozoa: why be normal? Microbes Infect. 2003;5:1231–1240. doi: 10.1016/j.micinf.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Palenchar JB, Bellofatto V. Gene transcription in trypanosomes. Mol. Biochem. Parasitol. 2006;146:135–141. doi: 10.1016/j.molbiopara.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Tschudi C, Ullu E. Unconventional rules of small nuclear RNA transcription and cap modification in trypanosomatids. Gene Expr. 2002;10:3–16. [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly S, Wickstead B, Gull K. An in silico analysis of trypanosomatid RNA polymerases: insights into their unusual transcription. Biochem. Soc. Trans. 2005;33:1435–1437. doi: 10.1042/BST0331435. [DOI] [PubMed] [Google Scholar]
- 20.Günzl A, Bruderer T, Laufer G, Schimanski B, Tu LC, Chung HM, Lee PT, Lee MG. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot. Cell. 2003;2:542–551. doi: 10.1128/EC.2.3.542-551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson PJ, Kooter JM, Borst P. Inactivation of transcription by UV irradiation of T. brucei provides evidence for a multicistronic transcription unit including a VSG gene. Cell. 1987;51:273–281. doi: 10.1016/0092-8674(87)90154-1. [DOI] [PubMed] [Google Scholar]
- 22.Sutton RE, Boothroyd JC. Evidence for trans splicing in trypanosomes. Cell. 1986;47:527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang XH, Haritan A, Uliel S, Michaeli S. Trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell. 2003;2:830–840. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakaar V, Dare AO, Hong D, Ullu E, Tschudi C. Upstream tRNA genes are essential for expression of small nuclear and cytoplasmic RNA genes in trypanosomes. Mol. Cell. Biol. 1994;14:6736–6742. doi: 10.1128/mcb.14.10.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chomczyinski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 26.Paillusson A, Hirschi N, Vallan C, Azzalin CM, Mühlemann O. A GFP-based reporter system to monitor nonsense-mediated mRNA decay. Nucleic Acids Res. 2005;33:e54. doi: 10.1093/nar/gni052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ullu E, Tschudi C. Permeable trypanosome cells as a model system for transcription and trans-splicing. Nucleic Acids Res. 1990;18:3319. doi: 10.1093/nar/18.11.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 29.Bochud-Allemann N, Schneider A. Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J. Biol. Chem. 2002;277:32849–32854. doi: 10.1074/jbc.M205776200. [DOI] [PubMed] [Google Scholar]
- 30.Devaux S, Lecordier L, Uzureau P, Walgraffe D, Dierick JF, Poelvoorde P, Pays E, Vanhamme L. Characterization of RNA polymerase II subunits of Trypanosoma brucei. Mol. Biochem. Parasitol. 2006;148:60–68. doi: 10.1016/j.molbiopara.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 31.Crausaz-Esseiva A, Marechal-Drouard L, Cosset A, Schneider A. The T-stem determines the cytosolic or mitochondrial localization of trypanosomal methionyl-tRNAs. Mol. Biol. Cell. 2004;15:2750–2757. doi: 10.1091/mbc.E03-11-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brun R, Schönenberger M. Cultivation an in vitro cloning of procyclic culture forms of Trypansoma brucei in a semi-defined medium. Acta Tropica. 1979;36:289–292. [PubMed] [Google Scholar]
- 33.Beverley SM, Clayton CE. Transfection of Leishmania and Trypanosoma brucei by electroporation. Methods Mol. Biol. 1993;21:333–348. doi: 10.1385/0-89603-239-6:333. [DOI] [PubMed] [Google Scholar]
- 34.Tan THP, Pach R, Crausaz A, Ivens A, Schneider A. tRNAs in Trypanosoma brucei: genomic organization, expression and mitochondrial import. Mol. Cell. Biol. 2002;22:3707–3717. doi: 10.1128/MCB.22.11.3707-3716.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pusnik M, Small I, Read LK, Fabbro T, Schneider A. Pentatricopeptide repeat proteins in Trypanosoma brucei function in mitochondrial ribosomes. Mol. Cell Biol. 2007;27:6876–6888. doi: 10.1128/MCB.00708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Domitrovich AM, Kunkel GR. Multiple, dispersed human U6 small nuclear RNA genes with varied transcriptional efficiencies. Nucleic Acids Res. 2003;31:2344–2352. doi: 10.1093/nar/gkg331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Padilla-Mejía NE, Florencio-Martínez LE, Figueroa-Angulo EE, Manning-Cela RG, Hernández-Rivas R, Myler PJ, Martínez-Calvillo S. Gene organization and sequence analyses of transfer RNA genes in Trypanosomatid parasites. BMC Genomics. 2009;10:232. doi: 10.1186/1471-2164-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 39.Siegel TN, Hekstra DR, Kemp LE, Figueiredo LM, Lowell JE, Fenyo D, Wang X, Dewell S, Cross GA. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009;23:1063–1076. doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bachellerie JP, Cavaillé J, Hüttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 41.Pusnik M, Charrière F, Mäser P, Waller RF, Dagley MJ, Lithgow T, Schneider A. The single mitochondrial porin of Trypanosoma brucei is the main metabolite transporter in the outer mitochondrial membrane. Mol. Biol. Evol. 2009;26:671–680. doi: 10.1093/molbev/msn288. [DOI] [PubMed] [Google Scholar]
- 42.Galli G, Hofstetter H, Birnstiel ML. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981;294:626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- 43.Xu XM, Carlson BA, Mix H, Zhang Y, Saira K, Glass RS, Berry MJ, Gladyshev VN, Hatfield DL. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007;5:e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hertz-Fowler C, Peacock CS, Wood V, Aslett M, Kerhornou A, Mooney P, Tivey A, Berriman M, Hall N, Rutherford K, et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–D343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krol ARvd, Mol JNM, Stuitje AR. Modulation of eukaryotic gene expression by complementary RNA or DNA sequences. Biotechniques. 1988;6:958–972. [PubMed] [Google Scholar]
- 46.Das A, Banday M, Bellofatto V. RNA polymerase transcription machinery in trypanosomes. Eukaryot. Cell. 2008;7:429–434. doi: 10.1128/EC.00297-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zamboni M, Scarabino D, Tocchini-Valentini GP. Splicing of mRNA mediated by tRNA sequences in mouse cells. RNA. 2009;15:2122–2128. doi: 10.1261/rna.1841609. [DOI] [PMC free article] [PubMed] [Google Scholar]