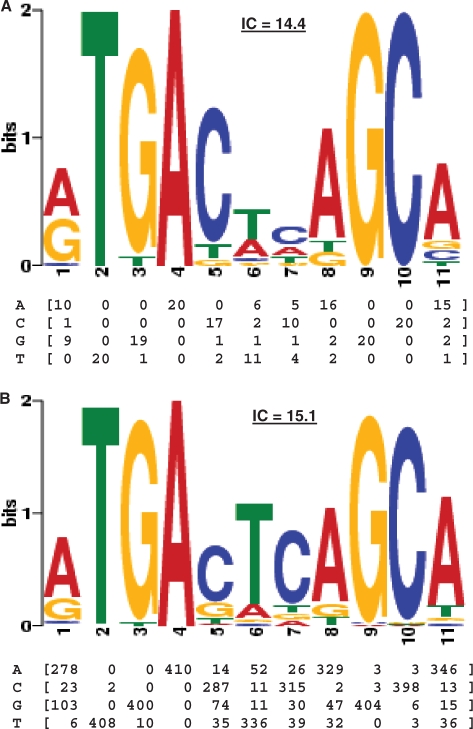
Figure 2.
Nrf2-binding profiles. (A) The original Nrf2-binding profile was defined by the alignment of the 20 known binding sites described in Table 1. (B) The motif generated by the MEME motif discovery algorithm (50) on the Nrf2 ChIP-Seq dataset is highly similar to the original profile with a higher information content (IC; in bits). The position Frequency Matrix (PFM) captures the count of each nucleotide found at each position in the motif and the logo provides a visualization of the data. The information content is the sum of the information content of each position and is computed as described previously (61).