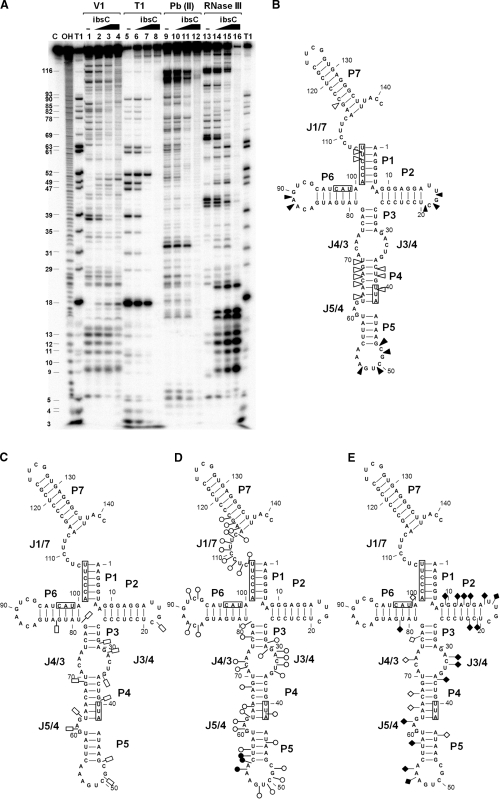
Figure 6.
Interaction of SibC with ibsC mRNA. (A) 32P-labeled SibC RNA (10 nM) was partially digested with RNase V1, RNase T1, lead(II) and RNase III in the absence (lanes 1, 5, 9 and 13) and presence of increasing amounts of unlabeled ibsC mRNA (25 nM final concentration: lanes 2, 6, 10 and 14; 100 nM final concentration: lanes 3, 7, 11 and 15). The fully paired SibC–ibsC mRNA complex was generated by heating and annealing with 100 nM ibsC. The complex was then used for digestion (lanes 4, 8, 12 and 16). Untreated SibC and alkaline ladders are shown in lanes C and OH, respectively. Lane T1 corresponds to the RNase T1 ladders of denatured SibC RNA. The positions of the cleaved G residues are shown on the left side of the gel. The altered sites of RNase V1 (B), RNase T1 (C), lead(II) (D) and RNase III (E) cleavage/protection in the presence of ibsC mRNA are shown in the secondary structure of SibC. The complementary sequences corresponding to the S/D region, start codon and stop codon of ibsC mRNA are indicated by boxes. Increasing and decreasing cleavages are indicated by solid and open symbols, respectively.