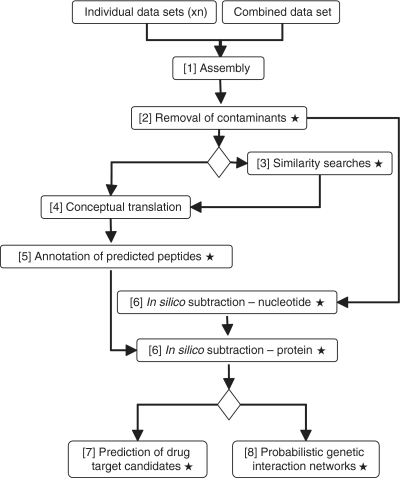
Figure 1.
Bioinformatic analyses of the Oesophagostomum dentatum data sets. Stars indicate analyses performed using custom-written Perl, Python and/or Unix shell computer scripts, accessible via http://research.vet.unimelb.edu.au/gasserlab/index.html. [1] Individual and combined expressed sequence tags (EST) data sets are assembled using CAP3 (compiled Linux 64-bit executable) to generate consensus sequences. [2] Assembled contigs with high similarity (cut-off: <1E-15) to nucleotide sequences of the vertebrate host (Sus scrofa) are eliminated. [3] Database similarity searches (for individual or combined data sets) are carried out using BLASTn and BLASTx (compiled Linux 64-bit executable; 42), embedded in custom-built Unix shell scripts. [4] Sequences (from the individually and combined assembled data sets) are conceptually translated into peptide sequences using ESTScan (compiled Linux 64-bit executable with a Perl wrapper). [5] Domains/motifs within translated peptides are identified via InterProScan (Perl wrapper) and linked to biological pathways in C. elegans using KOBAS (stand-alone Python application; 44). Functional annotation of the predicted peptides is performed by gene ontology (Perl wrapper; 27). [6] The individually assembled data sets are subtracted from one another (in both directions) using a BLASTn algorithm (42) embedded in a custom-built Unix shell script; proteins inferred from subtracted transcripts are assigned parental (i.e. level 1) InterPro terms and subtracted from one another using a BLASTp algorithm, embedded in a custom-built Unix shell script. [7] Potential drug target candidates for each of the individually assembled and/or in silico subtracted data sets are predicted and ranked according to the ‘severity’ of the non-wild-type RNAi phenotypes observed for the corresponding C. elegans orthologues/homologues (custom-built Unix shell scripts). [8] Probabilistic interaction networks among C. elegans orthologues of subtracted molecules are predicted (command lines).