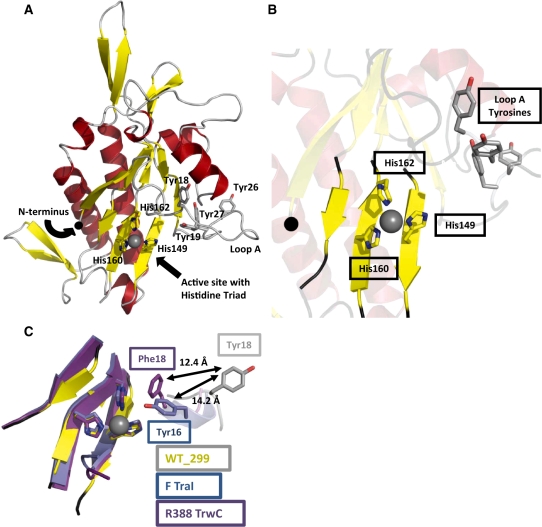
Figure 1.
The 2.3 Å resolution crystal structure of the pCU1 TraI relaxase. (A) The overall structure of the pCU1 relaxase (WT_299) with secondary-structures colored as represented in Figure 2 (β-strands are represented by yellow arrows, α-helices by red helices and loop regions by gray loops). The histidine triad of the active site and the tyrosines implicated in DNA nicking are shown as sticks. The manganese ion bound by the histidine triad is shown as a grey sphere. (B) The active site of the pCU1 relaxase (WT_299) with secondary structures represented as in (A). At the center is the histidine triad and bound manganese ion. In the upper right is loop A and the four tyrosines implicated in DNA nicking, and to the left is the N-terminus, represented as a black sphere. (C) The active site of the pCU1 relaxase [WT_299, colored as described in (A)] aligned with the active sites of the F plasmid TraI relaxase (in blue) and the relaxase of plasmid R388 TrwC (in magenta). The histidine triad and the primary DNA nicking tyrosine of each protein are shown as sticks. The distances between the tyrosines are indicated. Note that the in the structure of the plasmid R388 TrwC, Tyr18 was mutated to Phe18.