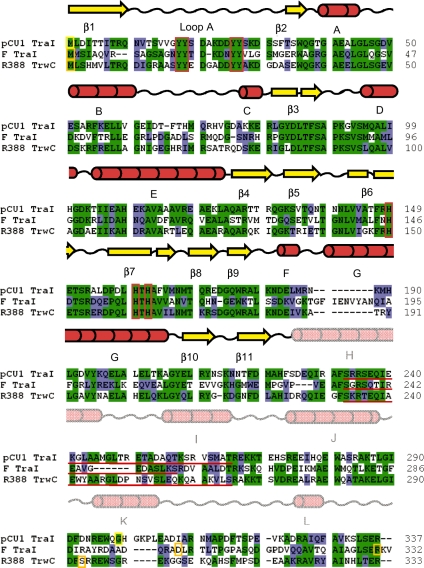
Figure 2.
Amino acid sequence alignment and secondary structure of the pCU1 TraI relaxase. The initial 337 amino acids of pCU1 TraI (GenBank ID AAD27542) were aligned with R388 TrwC (GenBank ID CAA44853) and F TraI (GenBank ID BAA97974) using the Clustal X program in BioEdit. Identical residues are shaded green, similar residues are shaded blue. Orange and yellow boxes indicate the first and last residues of the relaxase domains of each protein. Two constructs of the F TraI relaxase domain have been crystallized (3,23), therefore the terminal residues of both are boxed. Red boxes indicate the location of the conserved tyrosine residues and the active site histidine triad. Red lines underline the residues forming the ‘thumb’ regions of each protein. Since this region is disordered in the pCU1 TraI relaxase crystal, the extent of this region is estimated. Above the three sequences is the secondary structure of the pCU1 relaxase. β-strands are represented by yellow arrows; α-helices by red cylinders; and loop regions by black wavy lines. The secondary structure of residues 1–225 reflects that observed in the crystal structure of WT_299; the secondary structure of residues 226–337 reflects that predicted by Jpred 3.