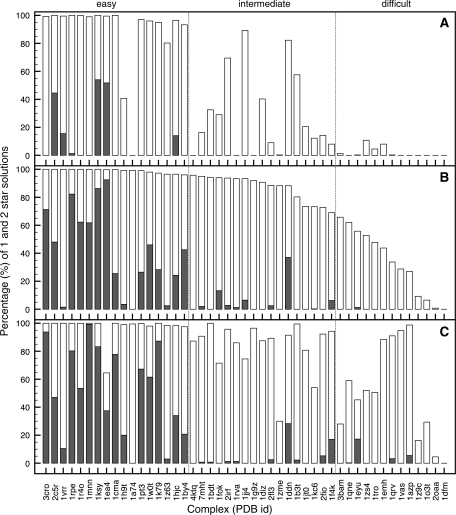
Figure 2.
Cumulative bar graphs expressing the quality of the best 400 docking solutions according to the HADDOCK score in terms of CAPRI one-star (grey) and two-star (white) results, for the two-stage unbound–unbound protein–DNA docking using true interface derived restraints. Results are presented for; the rigid-body docking starting from a canonical B-DNA model (A); after the semi-flexible refinement (B) and after semi-flexible refinement using an ensemble of custom DNA 3D structural models (C). Complexes are sorted according to the total number of obtained stars in (B), reclassifying the benchmark into ‘easy’, ‘intermediate’ and ‘difficult’ categories. See caption of Figure 1 for the definition of the CAPRI criteria.