FIG. 1.
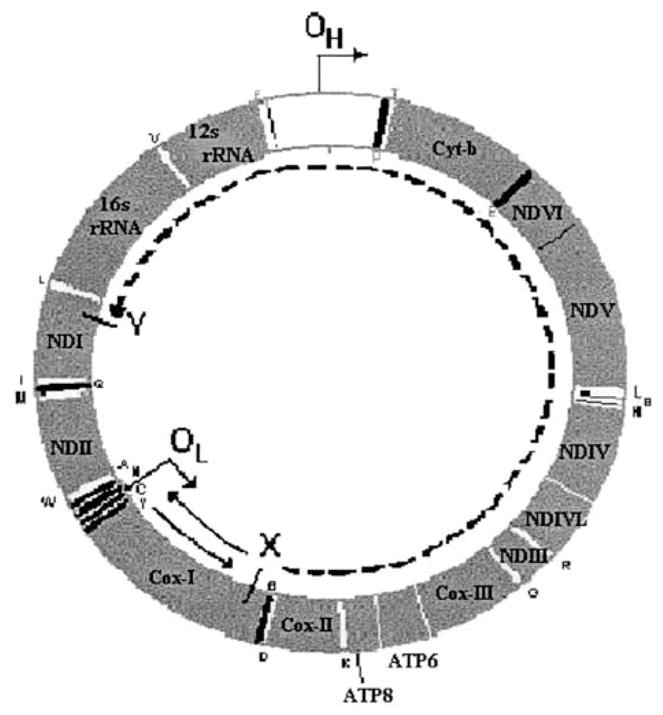
Time spent single stranded during replication of vertebrate mitochondria. The time that a site spends single stranded is determined by the site’s location. If a site is located after the origin of heavy-strand replication, OH (in the direction of heavy strand synthesis), but before the light-strand origin, OL (e.g., point X; this is case 1), then it will be single stranded for the time that it takes the heavy-strand replication fork to pass OL, plus the time for light-strand replication to begin, plus the time it takes for the light-strand replication fork to travel in the opposite direction and pass the site (e.g., return past point X). If one again assumes that initiation of light-strand synthesis is essentially instantaneous, and that the rate of movement of replication forks is constant, then the time that point X spends single stranded will be proportional to twice the distance from X to OL (solid lines.) If a site is past OL, however (e.g., point Y; case 2), then it will be single stranded for the time it takes for the light-strand replication fork to travel from wherever it is at that time to that point. If one assumes that the rate of movement of the replication forks is constant, then, for example, in the case of point Y, which is as distant from OL as is point X, then the time that point Y spends single stranded will be proportional to the distance from X to Y, in the direction of light-strand synthesis (dashed line). Since X and Y are equidistant from OL, this distance is proportional to the length of the genome minus twice the distance from X (or Y) to OL. The direction of replication from each of the origins is indicated by an arrow, protein-coding, and rRNA genes are labeled by their standard abbreviations, and tRNAs are labeled with their standard single-letter abbreviations.
