Figure 1.
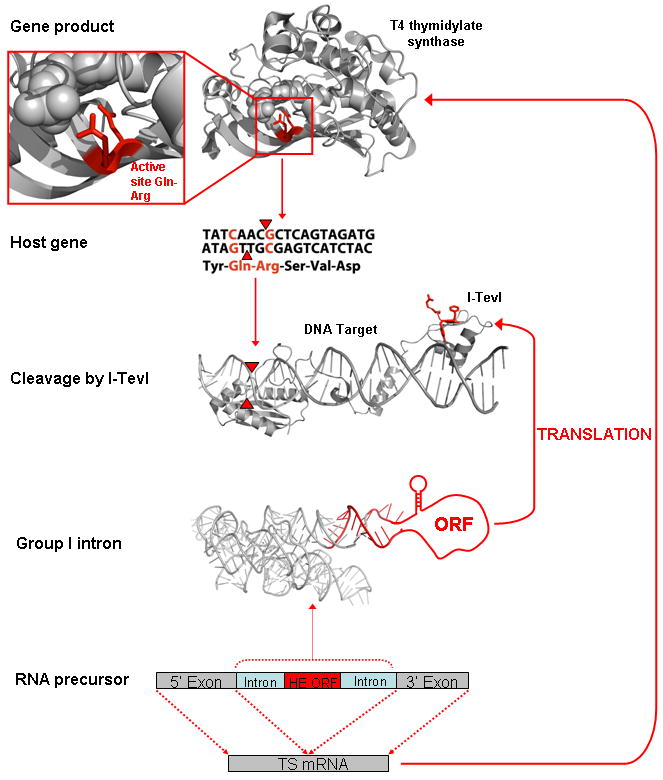
The mobile intron family circle. The RNA precursor of the interrupted T4 thymidylate synthase (TS) gene is depicted at the bottom of the figure, whereas the I-TevI intron-encoded homing endonuclease bound to its DNA target site in the T4 phage TS host gene is shown in the center. The endonuclease is highly specific for the presence of two conserved basepairs (red in host gene) in codons that specify two functionally critical conserved residues in the corresponding TS enzyme (top, Gln-Arg). These side chains contact the dUMP substrate (spheres in TS structure). Cleavage of the DNA target (red triangles) results in invasion of the host gene by the group I intron encoding I-TevI. The I-TevI ORF is inserted at a position corresponding to the P6 stem-loop and extends the final 18 bp at the 3′ end of the endonuclease ORF into the downstream intron core sequence (red bases extending beyond the intron P6 loop into P7). The amino acid residues in the endonuclease that are encoded by these basepairs (red side chains in I-TevI) are therefore constrained by the structure and function required both of the endonuclease and of the group I intron. The regulatory stem-loop, preceding the ORF in the group I intron, represses I-TevI translation, and its removal inhibits both splicing and TS synthesis. The depiction of the structure of full-length I-TevI and its DNA target site is a hypothetical, composite model composed of the crystal structure of its C-terminal domain bound to the 3′ end of its DNA target (pdb ID 1I3J) and a separate crystal structure of the unbound N-terminal catalytic domain (1LN0). The DNA target in the region of the nuclease domain, containing the points of DNA strand cleavage (red triangles) is modeled for illustrative purposes only.
