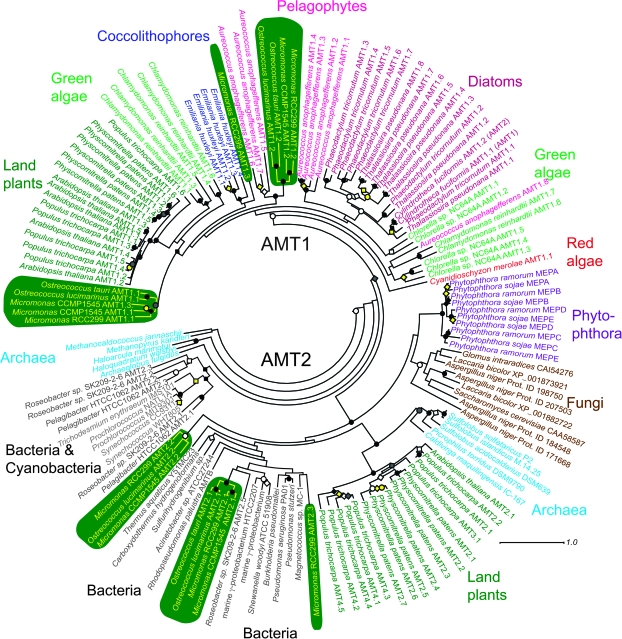
FIG. 2.
Unrooted maximum likelihood phylogenetic tree using protein sequences from AMT1 and AMT2 gene families from a selection of eukaryotes with sequenced genomes and from bacteria and archaea (some with sequenced genomes). Note that Chlamydomonas AMT1.2 is not shown but falls within Chlamydomonas subclass II. Transcripts of this gene have been cloned and sequenced, and the gene is present in the JGI v3 Chlamydomonas genome assembly, however, is missing from v4. Also AMT2B from O. tauri was not included because the 3' end of the gene sequence was truncated by a gap (in the genome sequence). Although S. cerevisiae has three MEPs, only one ScMEP is shown, although all three Aspergillus MEPs are included. Sequences at the most basal node (not shown) were AMT2.4 Roseobacter sp. SK209-2-6 (ZP_01755968), AMT2.4 Candidatus Pelagibacter ubique HTCC1062 (AAZ22114), and Silicibacter pomeroyi DSS-3 (YP_166819). Black symbols indicate bootstrap support between 95 and 100 (circles) or between 70 and 95 (diamonds) by both ML and NJ methods. White symbols represent the same for ML support only. Gray symbols represent the same for NJ methods only. Yellow symbols indicate bootstrap support between 95 and 100 (circles) for ML and 70 and 95 by NJ (circles) or 95 and 100 NJ and 70 and 95 ML support (diamonds). Accession numbers and gene model numbers are provided in supplementary table S5, Supplementary Material online.