Abstract
The Pdx1 or Ipf1 gene encodes an important homeodomain-containing protein with key roles in pancreas development and function. Mutations in human PDX1 are implicated in developmental defects and disease of the pancreas. Extensive research, including genome sequencing, has indicated that Pdx1 is the only member of its gene family in mammals, birds, amphibians, and ray-finned fish, and with the exception of teleost fish, this gene forms part of the ParaHox gene cluster along with Gsx1 and Cdx2. The ParaHox cluster, however, is a remnant of a 4-fold genome duplication; the three other ParaHox paralogues lack a Pdx-like gene in all vertebrate genomes examined to date. We have used bacterial artificial chromosome cloning and synteny analysis to show that the ancestor of living jawed vertebrates in fact had more ParaHox genes, including two Pdx genes (Pdx1 and Pdx2). Surprisingly, the two Pdx genes have been retained in parallel in two quite distantly related lineages, the cartilaginous fish (sharks, skates, and chimeras) and the Indonesian coelacanth, Latimeria menadoensis. The Pdx2 gene has been lost independently in ray-finned fish and in tetrapods.
Keywords: Pdx, Ipf, ParaHox, pancreas, coelacanth, Chondrichthyes
Introduction
One of the first vertebrate homeobox genes to be described was the Xenopus laevis XlHbox8 gene, expressed in a narrow band of endoderm in the embryo, with pancreatic expression in the adult frog (Wright et al. 1988). This was followed by the identification of the orthologous gene in mouse (insulin promoter factor 1 or Ipf1; Ohlsson et al. 1993) and rat (islet/duodenum homeobox 1 or idx1, also called somatostatin transactivating factor 1 or Stf1; Leonard et al. 1993; Miller et al. 1994). The mouse Ipf1 gene was mapped to distal chromosome 5 and the human orthologue to 13q12 (Fiedorek and Kay 1995; Stoffel et al. 1995). An orthologous gene was also described in zebrafish, medaka, and Xenopus tropicalis (Milewski et al. 1998; Assouline et al. 2002; Illes et al. 2009). The approved gene name according to the gene nomenclature committees for human and mouse is pancreatic and duodenal homeobox 1(Pdx1), and here, we utilize this nomenclature for the orthologous gene in all vertebrate species.
The vertebrate Pdx1 gene has a conserved role in the patterning of the midgut in all species examined to date. Mouse Pdx1 is expressed in the duodenum from around e8.5 and in the dorsal pancreatic bud from e12 (Jonsson et al. 1994). Homozygous Pdx1 mutants lack pancreatic tissue and parts of the posterior foregut, indicating an essential role for this gene in endodermal patterning in mouse (Jonsson et al. 1994; Offield et al. 1996). Similarly, pancreatic agenesis was reported in a human patient with a homozygous single-nucleotide deletion mutation, which resulted in formation of a truncated protein (Stoffers et al. 1997). Another patient with pancreatic agenesis was shown to be a compound heterozygote for two different nonsynonymous substitutions within the Pdx1 homeobox causing reduced protein half-life (Schwitzgebel et al. 2003).
In addition to its role in pancreas development, the Pdx1 protein is also a glucose-responsive regulator of the insulin gene in the β cells of the adult pancreas and is known to bind to the P1 enhancer 5′ of the insulin gene (Ohlsson et al. 1993). As a consequence of this role, mutations in Pdx1 have also been implicated in type II diabetes mellitus and maturity onset diabetes of the young type IV (Hani et al. 1999; Macfarlane et al. 1999; Cockburn et al. 2004).
The vertebrate Pdx1 gene was the only identified vertebrate member of the Xlox gene family, also called the Pdx gene family, which in turn is within the ANTP class of homeobox-containing genes (Wysocka-Diller et al. 1995; Holland et al. 2007). Members of the Xlox gene family are also present in many invertebrates; indeed, it was studies of the invertebrate amphioxus (Branchiostoma floridae) that revealed Xlox to be the central gene in an ancient cluster of three homeobox genes: Gsx, Xlox, and Cdx (Brooke et al. 1998). Comparison between many animal species indicates that this three-gene ParaHox cluster existed in the ancestor of all chordates and most likely existed in the ancestor of all bilaterian animals (Brooke et al. 1998; Ferrier and Holland 2001; Hui et al. 2009). Synteny analysis in the sea anemone Nematostella vectensis further suggests that a ParaHox cluster was present in the genome of the common ancestor of cnidarians and bilaterians, although it did not necessarily include an Xlox gene (Hui et al. 2008).
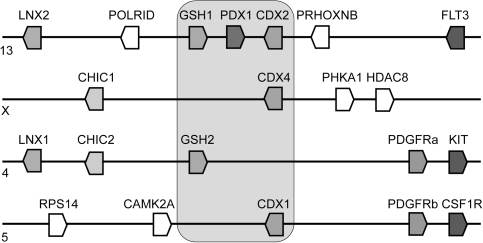
Although amphioxus has only a single ParaHox gene cluster, the ancestral cluster was duplicated 4-fold during early vertebrate evolution, as part of two whole-genome duplications (Coulier et al. 2000; Pollard and Holland 2000; Ferrier et al. 2005; Putnam et al. 2008). However, there was clearly extensive loss of duplicated genes because humans have only a single complete gene cluster comprising of one Gsx (GSH1), one Xlox (PDX1), and one Cdx (CDX2) on chromosome 13. The remaining duplicates each contain only a single ParaHox gene (GSH2 or CDX1 or CDX4), but analyses of flanking genes confirm that these four chromosomal regions are indeed descendent from the two genome duplications (fig. 1). In all vertebrates previously examined (human, mouse, chicken, frog, and several species of teleost fish), the complement of ParaHox genes is identical, with two Gsx genes (Gsx1 and Gsx2), the one Xlox gene (Pdx1), and three Cdx genes (Cdx1, Cdx2, and Cdx4). The situation is slightly complicated by an additional whole-genome duplication in teleost fish, but basically, the same complement is retained because teleost fish have lost cdx2 and instead have two copies of cdx1 (Mulley et al. 2006).
FIG. 1.
Genomic organization of ParaHox genes in the human genome. A cluster of Gsx, Xlox, and Cdx genes is present on chromosome 13. Paralogous genomic regions, generated by genome duplications, are found on chromosomes X, 4, and 5.
This consistency in gene complement is surprising because in early vertebrate evolution, there may have been up to four of each of these genes following the two rounds of whole-genome duplication. We therefore investigated the ParaHox gene complement of species at informative positions in the phylogeny of vertebrates: the Indonesian coelacanth (Latimeria menadoensis) as an outgroup to the tetrapods and three cartilaginous fish—lesser spotted dogfish (Scyliorhinus canicula), little skate (Leucoraja erinacea), and elephant shark (Callorhinchus milii)—as outgroups to bony vertebrates, such as ray-finned fish and tetrapods. We show that in both coelacanth and cartilaginous fish, there is an additional Pdx gene which we name Pdx2. Phylogenetic and phylogenomic analyses reveal that Pdx2 was present in the ancestor of all jawed vertebrates but has since been lost independently in both ray-finned fish and tetrapods.
Materials and Methods
Isolation of ParaHox Genes in Cartilaginous Fish and Coelacanths
Polymerase chain reaction (PCR) used genomic DNA of Indonesian coelacanth (L. menadoensis) and lesser spotted dogfish (S. canicula) and nested degenerate primers for the Xlox gene family (5′–3′: forward JMXloxIc: GACGACAACAAGMGNCANAGRAC; forward nested Xlox2: CAGCTGCTVGAGCTVGAGAA; reverse Xlox3: YTCCTCYTTYTTCCACTTCAT; reverse nested XSO2: GCGNCGRTTYTGGAACCAGAT), the Gsx gene family (forward JMGsx1a: ATGYCGMGVTCYTTYYWBGT; nested forward JMGsx1b: GTNGAYTCNYTVATNWTNARGGA; reverse Gsx3: TTGCCYTCYTTYTTGTGCTT; nested reverse GsxSO2: CANCKDCGRTTYTGRAACCA), and the Cdx gene family (5'–3': forward JmCdx: GGNAARCANMGRACVAARGA; nested forward CdxSO1: CTRGARCTGGARAARGARTT; reverse CdxSO2: NVKNVKRTTYTGRAACCA). Rapid amplification of cDNA ends polymerase chain reaction in S. canicula used the SMART RACE cDNA Amplification Kit (Clontech 634914) and pooled embryonic cDNA as template. Full coding sequences of S. canicula Pdx1 and Pdx2 are deposited in GenBank under the accession numbers HM142925 and HM142926. Blast searches of the elephant shark C. milii partial genome sequence (Venkatesh et al. 2007) were carried out using the Blast server on the Elephant Shark Genome Project homepage (http://esharkgenome.imcb.a-star.edu.sg/). In order to analyze the genomic location of Pdx2, we screened high coverage bacterial artificial chromosome (BAC) libraries for Indonesian coelacanth (from the Genome Resource Centre, Benaroya Research Institute, Seattle, WA) and little skate L. erinacea (from Clemson University Genomics Institute, Clemson, SC) using digoxigenin-labeled Pdx1 and Pdx2 homeobox probes. Positive clones were verified by PCR using the above primers prior to sequencing. Coelacanth BAC clone 188I4 containing Pdx2 was sequenced to 9.7× coverage using Sanger sequencing (performed at the Washington University Genome Centre, St Louis, MI); little skate BAC clone 24D8 was sequenced to 45× coverage using Roche 454 GS FLX Titanium technology (performed at the Centre for Genomic Research, University of Liverpool, UK). Genes were predicted using Blast and GenScan and by alignment to known orthologous genes. BAC clone sequences are deposited in GenBank under accession numbers HM134894 and HM134895.
Phylogenetic Analysis
Orthology and paralogy of Gsx, Pdx, and Pdgfr genes in sequenced BAC clones and related loci in other vertebrates were resolved using phylogenetic analysis. Amino acid sequences were aligned using ClustalX (Larkin et al. 2007) and edited by eye to maximize contiguity of alignable sequence; maximum likelihood phylogenetic trees were constructed with PhyML (Guindon and Gascuel 2003) using the JTT matrix and 1,000 bootstrap replicates. The resulting trees are provided as supplemental information (Supplementary Material online).
Gene Expression Analysis
Expression of Pdx1 and Pdx2 in adult dogfish tissues was analyzed using reverse transcription--polymerase chain reaction (RT–PCR). Total RNA was extracted using TRI Reagent (Applied Biosystems Inc., AM9738) in accordance with the supplier’s instructions and treated with RNase-free DNaseI (New England Biolabs, M0303) to remove contaminating genomic DNA. Single-stranded cDNA was synthesized using Bioline cDNA Synthesis Kit (BIO-65025) with oligo (dT) priming and PCR carried out using primers ScPdx1f1: AGAGGATCCTACCGTCTCGCATC; ScPdx1r1: CACCGAGTCTCTCGTAGCCGTAG; ScPdx2f1: ACGGATTTCACTGGCTACGACAC; ScPdx2r1: ACCAGATTTTGATGTGCCTCTCG; ScActinf1: AGTTGGATGGGTCAGAAAGAC; and ScActinR1: ACGCTCAGTCAGGATCTTCATC.
Results
All jawed vertebrates examined previously were found to have an equivalent complement of ParaHox genes, comprising two Gsx genes, one Xlox gene, and three Cdx genes. We asked whether this was the ancestral condition for jawed vertebrates. Key unrepresented informative positions in the evolutionary tree of vertebrates are the coelacanths, representing an early diverging branch within the sarcopterygians, and the sharks, skates, and holocephalians, representing the chondrichthyans. To examine the complement of ParaHox genes in these groups, we first used degenerate PCR on genomic DNA from the Indonesian coelacanth (L. menadoensis) and the lesser spotted dogfish (S. canicula), plus analysis of partial genome sequence information from a holocephalan, the elephant shark (C. milii). The most complete data were derived from the dogfish, which had the expected complement of Gsx1, Gsx2, Pdx1, Cdx1, Cdx2, and Cdx4 genes, whereas fragments of all but Cdx2 were found in the available elephant shark sequence. These analyses also revealed the presence of an extra homeobox sequence assignable to the Xlox family, in addition to the expected Pdx1 gene. The new locus, which we call Pdx2, has not been reported in any other vertebrate species examined to date (fig. 2). The full coding sequence of the dogfish Pdx2 gene was obtained by RACE PCR using pooled embryonic cDNA.
FIG. 2.
Amino acid alignment of Xlox homeodomains. Only cartilaginous fish and coelacanth have two members of the Xlox gene family—Pdx1 and Pdx2. Pdx2 sequences shown here were derived from BAC clones (coelacanth and little skate), RACE PCR (dogfish), and genomic sequence (elephant shark).
There are three alternative hypotheses that could account for the presence of the additional Pdx genes: independent lineage-specific gene duplication in the coelacanth and chondrichthyan lineages, parallel retention of an ancestral vertebrate Pdx2 gene with independent loss in actinopterygians and tetrapods, or duplication in one lineage (coelacanths or chondrichthyans) with retention of an ancestral gene in the other (chondrichthyans or coelacanths). To resolve between these hypotheses, we first constructed phylogenetic trees to test for orthology. These suggested that the Pdx2 homeobox sequences of coelacanth and cartilaginous fish form a monophyletic group and are likely to be orthologues rather than independent duplications in each lineage (supplementary information, Supplementary Material online). However, these initial analyses used only a short sequence length (95 aa) and are not conclusive. To provide a definitive test, we needed to determine the genomic location of the Pdx2 loci, particularly in relation to their neighboring genes. These could then be compared with the well-studied paralogy groups around the mammalian and teleost ParaHox genes.
Unfortunately, as yet, there is no assembled genome sequence for coelacanth (although a genome project has been approved by the National Human Genome Research Institute) and available sequences from the Elephant Shark Genome Project do not assemble into large contigs, precluding analysis of the Pdx2 gene neighbors. We therefore screened large insert BAC libraries from coelacanth and little skate (the latter species being closely related to lesser spotted dogfish for which there is no high coverage BAC library).
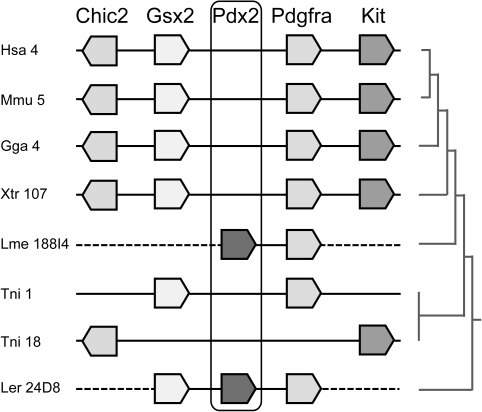
A Pdx2-positive BAC clone was identified in each species and sequenced. First, this allowed us to extend the deduced coding sequence for Pdx2 from a coelacanth and a chondrichthyan; alignment of these revealed close similarity, further supporting orthology between the two (data not shown). More importantly, the BAC sequencing revealed that the Pdx2 gene of coelacanth is linked to the platelet-derived growth factor receptor α (Pdgfrα) gene, and the Pdx2 gene of little skate is linked to the Gsx2 homeobox gene and Pdgfrα gene (fig. 3). In each case, the orthology of the neighbor gene was verified by phylogenetic analysis (supplementary information, Supplementary Material online). These findings demonstrate that Pdx2 in both coelacanths and chondrichthyans is located in the ParaHox paralogy region equivalent to that present at human chromosomal location 4q12 (figs. 1 and 3). Indeed, analysis of the orthologous region in teleost fish and tetrapods shows that Gsx2 is usually located next to Pdgfrα, with no intervening genes (fig. 3). It is well established that the four ParaHox paralogy regions were generated by two rounds of whole-genome duplication early in vertebrate ancestry. Hence, Pdx2 must be an ancient vertebrate gene that was generated during these genome duplication events and was subsequently lost independently in teleosts and tetrapods but retained in parallel in coelacanths and chondrichthyans (fig. 4).
FIG. 3.
Genomic neighborhood of the Gsx2 locus in vertebrates. Coelacanth and little skate have a Pdx2 gene located between Gsx2 and Pdgfrα. Hsa, Homo sapiens; Mmu, Mus musculus; Gga, Gallus gallus; Xtr, Xenopus tropicalis; Lme, Latimeria menadoensis; Tni, Tetraodon nigroviridis; Ler, Leucoraja erinacea; numbers refer to chromosome number or genomic scaffold.
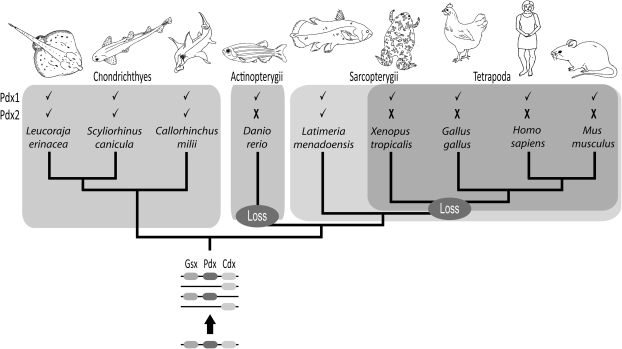
FIG. 4.
Evolutionary history of ParaHox genes in vertebrates. The vertebrate ancestor had a Pdx2 gene linked to Gsx2 in a ParaHox paralogon; this gene has been lost independently in ray-finned fish and tetrapods and retained in parallel in cartilaginous fish and coelacanths.
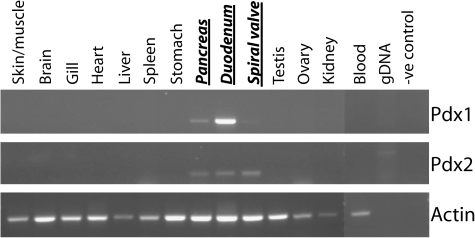
The parallel retention of an ancient homeobox gene in two widely divergent evolutionary lineages raises intriguing questions about the roles of the Pdx1 and Pdx2 genes. In order to shed light on this subject, we carried out RT–PCR using RNA extracted from adult dogfish tissues. This revealed that the Pdx1 gene is expressed most strongly in the pancreas and associated duodenum, with weak expression in the spiral valve. The Pdx2 gene shows a similar expression profile and therefore overlaps with Pdx1, although Pdx2 is expressed more highly in the spiral valve (fig. 5). Analyses of gene expression in coelacanths are not currently possible because of their rarity and protection under Appendix I of the Convention on International Trade in Endangered Species.
FIG. 5.
Expression of Pdx1 and Pdx2 in adult dogfish tissues. RT–PCR for both Pdx genes in a number of different tissues reveals that Pdx1 and Pdx2 expression is restricted to the pancreas and duodenum (including the spiral valve).
Discussion
Homeobox genes have provided important insights into vertebrate genome evolution and gene family diversification. We have investigated the pattern of evolution of the developmentally and medically important ParaHox gene family in vertebrates using species chosen for their phylogenetic positions. One question asked was why is there an apparently stable complement of ParaHox genes in jawed vertebrates, comprising two Gsx genes, one Xlox gene (Pdx1), and three Cdx genes? Here, we show that in fact, this complement is not stable nor is it retained in all vertebrate lineages. We have identified a previously unknown member of the Xlox gene family (Pdx2) and, by analyses of its genomic context, discovered that this gene was present in the ancestor of all living jawed vertebrates. Intriguingly, however, Pdx2 has been retained during vertebrate evolution only in cartilaginous fish and coelacanths (fig. 4), two quite distant lineages, and has been lost independently in ray-finned fish and tetrapods.
The retention of both Pdx1 and Pdx2 genes in any living vertebrate indicates that the two genes cannot have completely equivalent roles; otherwise, one gene would have been lost by mutation and degeneration in the 450–550 My since their origin. The earliest stage of this functional distinction between Pdx1 and Pdx2 is likely to have occurred at the gene regulatory level, possibly involving loss of ancestral (preduplication) roles in each of the daughter genes in a complementary manner (subfunctionalization; Lynch and Force 2000). It may have taken tens of millions of years for these regulatory mutations to accumulate sufficiently for gene expression divergence, judging by evidence from other gene families, such as neurogenin (Furlong and Graham 2005) and snail (Locascio et al. 2002). For example, mammals and teleost fish express neurogenin 1 (Ngn1) in the ophthalmic trigeminal placode where birds use neurogenin 2 (Ngn2); this implies that Ngn1 and Ngn2 were both expressed in this placode prior to the divergence of birds and mammals, with their differential expression evolving long after the actual duplication event (Furlong and Graham 2005). If a similar process took place with vertebrate Pdx1 and Pdx2, then for a substantial period of time after their origin they will have had identical roles, followed by stepwise subfunctionalization over millions of years and possibly addition of new functions. However, even though there must be functional differences between Pdx1 and Pdx2 in living vertebrates, our analyses of gene expression in dogfish have not thus far revealed any clear distinction between the two genes, with both being expressed in pancreas, duodenum, and the spiral valve. Because the subfunctionalizing mutations will have occurred independently in chondrichthyans and in coelacanths, we should not assume that the two genes are performing exactly the same role in these two evolutionary lineages.
It is clear that the Pdx1 and Pdx2 genes were generated in the genome duplication events that occurred during early vertebrate evolution and that both were retained when the jawed vertebrate lineage split into the Chondrichthyes (cartilaginous fish) and Osteichthyes (bony vertebrates). In the lineage leading to humans, the Osteichthyes, the Pdx2 gene was retained until the divergence of Actinopteryggii and the Sarcopterygii, and in latter lineage, it was still present at the divergence of the coelacanth and tetrapod lineages. The gene was then lost independently within the Actinopterygii and in the tetrapods, before the evolutionary radiation of the living tetrapod lineage. Lungfish are the only group of animals likely to have diverged in this period, being the probable sister group to tetrapods (Takezaki et al. 2003). In an attempt to refine the timing of Pdx2 gene loss in sarcopterygians, we used PCR to search for Pdx genes in the Australian lungfish (Neoceratodus forsteri). We identified only the Pdx1 gene, tentatively suggesting that Pdx2 may have been lost in the ancestor of lungfish and tetrapods (data not shown). As to why Pdx2 was lost twice in vertebrate evolution, in tetrapods and in teleosts, it is key to recall the process of duplicate gene divergence discussed above. If functional divergence occurred over a period of tens of millions of years, then the two genes could still have had substantially overlapping roles for the entire time between the gene duplication event and the divergence of Actinoptergygii and the Sarcopterygii. This period may have been less than 100 My, if we consider the genome duplication date to be 450–550 Ma and the Actinoptergygii/Sarcoptergii divergence as 425–476 Ma (Blair and Hedges 2005). After that phylogenetic node, we deduce that disabling mutations occurred in Pdx2 independently in a teleost ancestor and a tetrapod ancestor and were viable because of redundancy with Pdx1. We assume that it would have been possible to lose either one of the genes and still maintain full function, and thus, it is coincidence that both teleosts and tetrapods both lost Pdx2 rather than Pdx1. An alternative explanation is that the full function could only be fulfilled by Pdx1, perhaps because of some feature of its genomic location, but we find no evidence to support this suggestion because both Pdx genes are linked to Gsx genes, whereas teleost fish genomes demonstrate that Pdx1 can function without a neighboring Cdx2 gene. Either way, the independent loss of Pdx2 in teleosts and tetrapods, and its parallel retention in chondrichthyans and coelacanths, presents a particularly unusual pathway of molecular evolution for vertebrate homeobox genes.
Supplementary Material
Supplementary materials are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
This research was supported by a grant from the Wellcome Trust (081233/Z/06/Z). We thank Harv Isaacs for useful discussions, Ashley Tweedale and David Sims for supplying dogfish material, Chris Amemiya and Axel Meyer for coelacanth material, and Anjana Badrinarayanan for assistance with initial PCR experiments. The authors also acknowledge the artistic skills of Tatiana Solovieva for help with the figures.
References
- Assouline B, Nguyen V, Mahe M, Bourrat F, Scharfmann R. Development of the pancreas in medaka. Mech Dev. 2002;117:299–303. doi: 10.1016/s0925-4773(02)00190-9. [DOI] [PubMed] [Google Scholar]
- Blair JE, Hedges SB. Molecular phylogeny and divergence times of deuterostome animals. Mol Bio Evol. 2005;22:2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- Brooke NM, Garcia-Fernàndez J, Holland PWH. The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature. 1998;392:920–922. doi: 10.1038/31933. [DOI] [PubMed] [Google Scholar]
- Cockburn BN, Bermano G, Boodram L-LG, Teelucksingh S, Tsuchiya T, Mahabir D, Allan AB, Stein R, Docherty K, Bell GI. Insulin promoter factor-1 mutations and diabetes in Trinidad: identification of a novel diabetes-associated mutation (E224K) in an Indo-Trinidadian family. J Clin Endocrinol Metab. 2004;89:971–978. doi: 10.1210/jc.2003-031282. [DOI] [PubMed] [Google Scholar]
- Coulier F, Burtey S, Chaffanet M, Birg F, Birnbaum D. Ancestrally duplicated ParaHox gene clusters in humans. Int J Oncol. 2000;17:439–444. doi: 10.3892/ijo.17.3.439. [DOI] [PubMed] [Google Scholar]
- Ferrier DEK, Dewar K, Cook A, Chang JL, Hill-Force A, Amemiya C. The chordate ParaHox cluster. Curr Biol. 2005;15:R820–R822. doi: 10.1016/j.cub.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Ferrier DEK, Holland PWH. Sipunculan ParaHox genes. Evol Dev. 2001;3:263–270. doi: 10.1046/j.1525-142x.2001.003004263.x. [DOI] [PubMed] [Google Scholar]
- Fiedorek FT, Kay ES. Mapping of the insulin promoter factor 1 gene to distal mouse chromosome 5. Genomics. 1995;28:581–584. doi: 10.1006/geno.1995.1193. [DOI] [PubMed] [Google Scholar]
- Furlong RF, Graham A. Vertebrate neurogenin evolution: long-term maintenance of redundant duplicates. Dev Genes Evol. 2005;215:639–644. doi: 10.1007/s00427-005-0023-x. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hani EH, Stoffers DA, Chevre J-C, Durand E, Stanojevic V, Dina C, Habener JF, Froguel P. Defective mutations in the insulin promoter factor-1 (IPF-1) gene in late-onset type 2 diabetes mellitus. J Clin Invest. 1999;104:R41–R48. doi: 10.1172/JCI7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PWH, Booth HAF, Bruford E. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007;5:47. doi: 10.1186/1741-7007-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui JH, Holland PWH, Ferrier DEK. Do cnidarians have a ParaHox cluster? Analysis of synteny around a Nematostella homeobox gene cluster. Evol Dev. 2008;10:725–730. doi: 10.1111/j.1525-142X.2008.00286.x. [DOI] [PubMed] [Google Scholar]
- Hui JH, Raible F, Korchagina N, et al. (11 co-authors) Features of the ancestral bilaterian inferred from Platynereis dumerilii ParaHox genes. BMC Biol. 2009;7:43. doi: 10.1186/1741-7007-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illes JC, Winterbottom E, Isaacs HV. Cloning and expression analysis of the anterior parahox genes, Gsh1 and Gsh2 from Xenopus tropicalis. Dev Dyn. 2009;238:194–203. doi: 10.1002/dvdy.21816. [DOI] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin promoter factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. (13 co-authors) Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Leonard JB, Peers T, Johnson K, Ferrere S, Lee S, Montminy M. Characterization of somatostatin transactivating factor-1, a novel homeobox factor that stimulates somatostatin expression in pancreatic islet cells. Mol Endocrinol. 1993;7:1275–1283. doi: 10.1210/mend.7.10.7505393. [DOI] [PubMed] [Google Scholar]
- Locascio A, Manzanares M, Blanco MJ, Nieto MA. Modularity and reshuffling of Snail and Slug expression during vertebrate evolution. Proc Natl Acad Sci U S A. 2002;99:16841–16846. doi: 10.1073/pnas.262525399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane WM, Frayling TM, Ellard S, et al. (14 co-authors) Missense mutations in the insulin promoter factor-1 gene predispose to type 2 diabetes. J Clin Invest. 1999;104:R33–R39. doi: 10.1172/JCI7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewski WM, Duguay SJ, Chan SJ, Steiner DF. Conservation of PDX-1 structure, function, and expression in zebrafish. Endocrinology. 1998;139:1440–1449. doi: 10.1210/endo.139.3.5768. [DOI] [PubMed] [Google Scholar]
- Miller CP, McGehee RE, Habener JF. IDX-1: a new homeodomain transcription factor expressed in rat pancreatic islets and duodenum that transactivates the somatostatin gene. EMBO J. 1994;13:1145–1156. doi: 10.1002/j.1460-2075.1994.tb06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulley JF, Chiu C-H, Holland PWH. Break-up of a homeobox gene cluster after genome duplication in teleosts. Proc Natl Acad Sci U S A. 2006;103:10369–10372. doi: 10.1073/pnas.0600341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CVE. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Ohlsson H, Karlsson K, Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 1993;12:4251–4259. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard SL, Holland PWH. Evidence for 14 homeobox gene clusters in human genome ancestry. Curr Biol. 2000;10:1059–1062. doi: 10.1016/s0960-9822(00)00676-x. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DEK, et al. (36 co-authors) The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1072. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Schwitzgebel VM, Mamin A, Brun T, et al. (13 co-authors) Agenesis of human pancreas due to decreased half-life of insulin promoter factor 1. J Clin Endocrinol Metab. 2003;88:4398–4406. doi: 10.1210/jc.2003-030046. [DOI] [PubMed] [Google Scholar]
- Stoffel M, Stein R, Wright CVE, Espinosa R, Le Beau MM, Bell GI. Localization of human homeodomain transcription factor Insulin Promoter Factor 1 (IPF1) to chromosome band 13q12.1. Genomics. 1995;28:125–126. doi: 10.1006/geno.1995.1120. [DOI] [PubMed] [Google Scholar]
- Stoffers DA, Zinkin NT, Stanojevic V, Clarke WL, Habener JF. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- Takezaki N, Figueroa F, Zaleska-Rutczynska Z, Klein J. Molecular phylogeny of early vertebrates: monophyly of the agnathans as revealed by sequences of 35 genes. Mol Biol Evol. 2003;20:287–292. doi: 10.1093/molbev/msg040. [DOI] [PubMed] [Google Scholar]
- Venkatesh B, Kirkness EF, Loh YH, et al. (12 co-authors) Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol. 2007;5:e101. doi: 10.1371/journal.pbio.0050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CVE, Schnegelsberg P, De Robertis EM. XlHbox8: a novel Xenopus homeo protein restricted to a narrow band of endoderm. Development. 1988;104:787–794. doi: 10.1242/dev.105.4.787. [DOI] [PubMed] [Google Scholar]
- Wysocka-Diller J, Aisemberg GO, Macango ER. A novel homeobox cluster expressed in repeated structures of the midgut. Dev Biol. 1995;171:439–447. doi: 10.1006/dbio.1995.1294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.