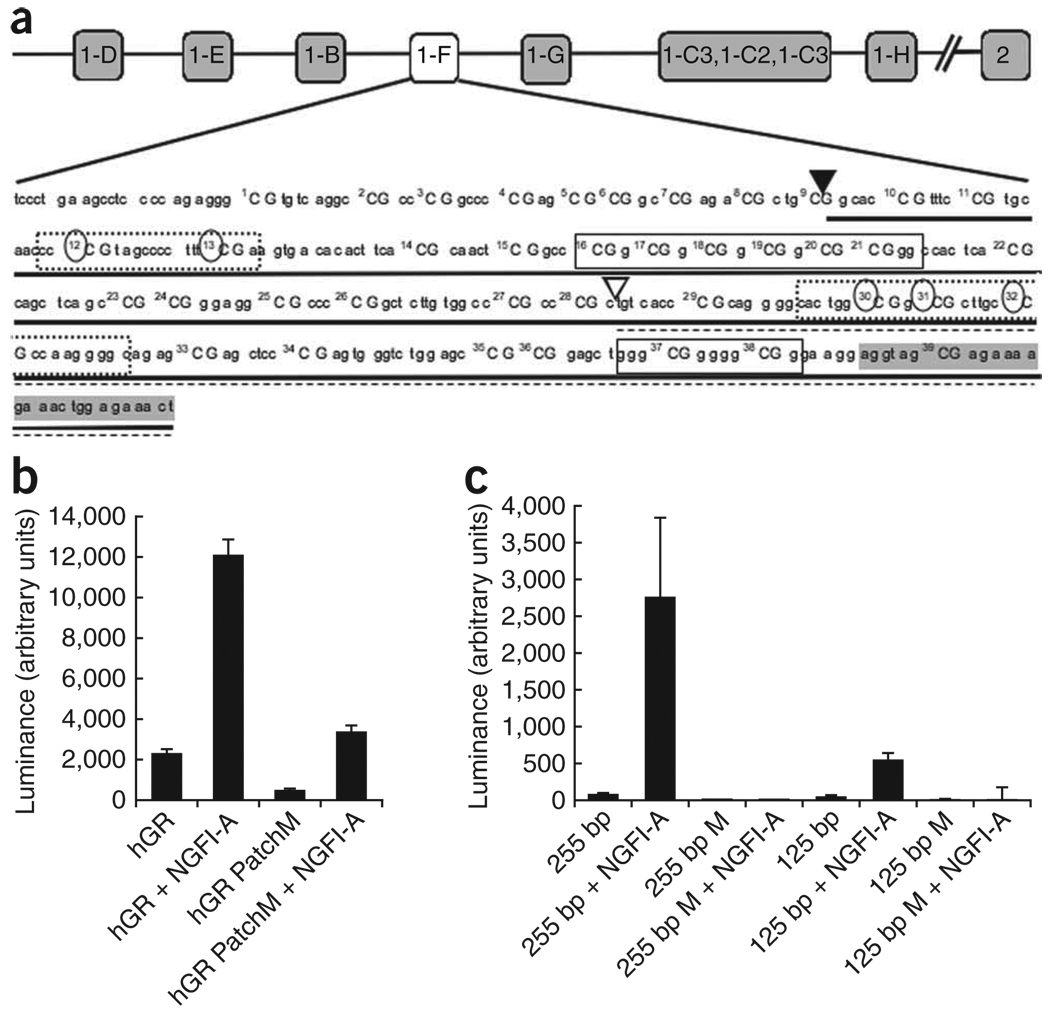
Figure 3.
In vitro analysis of NR3C1 promoter methylation. (a) The relative position of the NR3C1 variant and the promoter sequence, showing the location of the CpG dinucleotides. The 255-bp (▼, solid underline) and 125-bp (∇, broken underline) deletion constructs are shown, along with specific CpG dinucleotides that were methylated in each deletion construct, as indicated by circles. Boxes represent known or putative canonical (solid-lined box) and noncanonical (broken-lined box) NGFI-A–binding sites, with the shaded area indicating the beginning of the exon. (b,c) Mean ± s.e.m. levels of luciferase expression in HEK293 cells. Results are shown after the subtraction of expression of the promoter in the antisense orientation. (b) The full NR3C1 promoter was either unmethylated (hGR) or completely patch methylated (hGR PatchM) and transfected in the presence or absence of NGFI-A. (c) The 255-bp and 125-bp NR3C1 deletion construct were either unmethylated (255 bp or 125 bp) or methylated (255 bp M or 125 bp M), as shown in a, and transfected either in the presence or absence of NGFI-A.