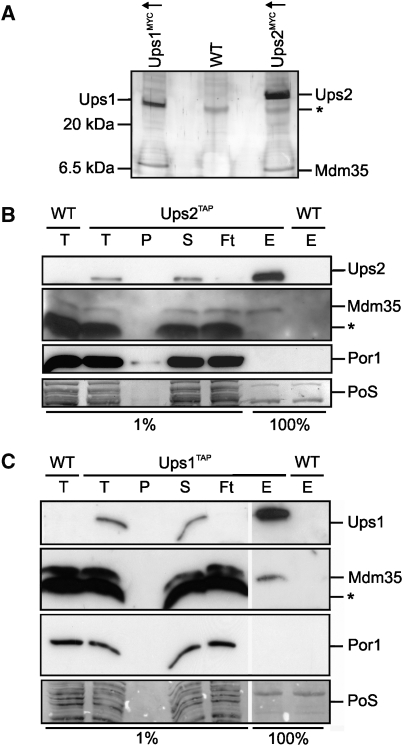
Figure 1.
Mdm35 interacts with Ups1 and Ups2. (A) Coimmunoprecipitation of Mdm35 with Ups1MYCand Ups2MYC. Mitochondria isolated from wild-type cells (WT, CG214) or cells overexpressing either Ups1MYC (Ups1MYC↑, CG630) or Ups2MYC (Ups2MYC↑, CG626) were solubilized and subjected to coimmunoprecipitation using MYC-specific antibodies. Eluates were analysed by Tris/Tricine SDS–PAGE and subsequent silver staining. Ups1, Ups2 and Mdm35 were identified by peptide mass finger printing. The asterisk (*) marks a protein binding unspecifically to the beads. (B, C) Affinity purification of Mdm35 with endogenous levels of Ups1TAP and Ups2TAP. Mitochondria isolated from wild-type cells (WT,CG214) or cells expressing either (B) Ups2TAP (CG591) or (C) Ups1TAP (CG593) from the endogenous gene locus were solubilized and proteins were affinity purified using immunoglobulin G-coupled beads. Total (T), pellet (P), supernatant (S) flow-through (Ft) fractions (1%) and the eluate (E) fraction (100%) were analysed by Tris/Tricine SDS–PAGE, followed by Ponceau S staining (PoS) and immunoblotting. The asterisk (*) marks a protein band cross-reacting with Mdm35 antibodies.