Figure 4.
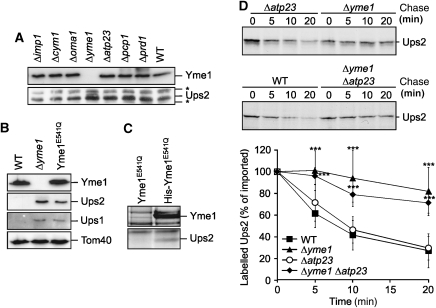
Ups2 is degraded by the i-AAA protease Yme1. (A) Accumulation of Ups2 in Δyme1 cells. Wild-type cells (WT, CG214) and cells lacking different mitochondrial peptidases [Δimp1 (Euroscarf 732), Δcym1 (Euroscarf 4266), Δoma1 (Euroscarf 6003), Δyme1(CG113), Δatp23 (CW1), Δpcp1 (Euroscarf 4731), Δprd1(Euroscarf 3464)] were grown to mid-log phase on glucose-containing media. Mitochondria-enriched membrane fractions were analysed by SDS–PAGE and immunoblotting using Yme1- and affinity-purified Ups2-specific antibodies. Asterisks (*) indicate protein bands cross-reacting with the Ups2-antibodies. (B) Ups1 and Ups2 accumulate in mitochondria lacking proteolytically active Yme1. Mitochondria isolated from wild-type cells (WT, CG1), Δyme1(VIA4) cells or Δyme1 cells expressing proteolytically inactive Yme1E541Q (YTE26) were analysed by SDS–PAGE and immunoblotting using Yme1-, Tom40- and affinity-purified Ups1- and Ups2-specific antibodies. (C) Binding of Ups2 to proteolytically inactive Yme1E541Q. Mitochondria isolated from yme1 cells expressing either Yme1E541Q (YTE26) or a variant thereof carrying an N-terminal hexahistidine tag (His Yme1E541Q) (YTE89) were solubilized and extracts subjected to Ni-NTA affinity chromatography. Eluate fractions were analysed by SDS–PAGE and Coomassie staining. Ups2 was detected by peptide mass finger printing in the eluate only when mitochondria harbouring His-Yme1E541Q were analysed. (D) Degradation of Ups2 by Yme1 following import into mitochondria in vitro. Radiolabelled Ups2 was imported into mitochondria derived from either wild-type (WT, CG1), Δyme1 (VIA4), Δatp23 (CW3) or Δyme1Δatp23 (CW79) cells. The stability of newly imported Ups2 upon further incubation at 37°C was assessed by SDS–PAGE and autoradiography. Quantification of Ups2 accumulation in mitochondria is represented in the lower panel. Newly imported Ups2 was set to 100%. Data represent ±s.d. of four independent experiments. ***P<0.0005.