Figure 6.
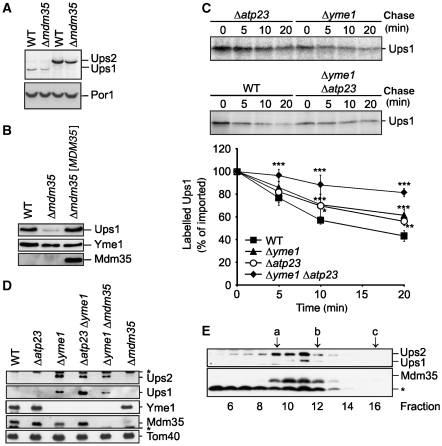
Mdm35 controls degradation of Ups1 by Yme1 and Atp23. (A) Ups1 is less abundant than Ups2. Wild-type (WT) and Δmdm35 cells expressing genomically tagged Ups2MYC (PY102, PY117) or Ups1MYC (PY103, CW385) were grown to mid-log phase. Proteins were extracted and analysed by SDS–PAGE and immunoblotting using MYC- and porin (Por1)-specific antibodies. (B) Steady-state levels of Ups1MYC in the absence of Mdm35. Wild-type (WT) and Δmdm35 cells expressing genomically modified Ups1MYC were transformed with either YCplac111∷ADH (CW394, CW398) or YCplac111∷ADH encoding MDM35 ([MDM35]) (CW396). Mitochondria-enriched membrane fractions were analysed by SDS–PAGE and immunoblotting using MYC-, Yme1- and affinity-purified Mdm35-specific antisera. (C) Ups1 is stabilized in mitochondria lacking YME1 and ATP23. Radiolabelled Ups1 was imported into mitochondria isolated from WT (CG1), Δyme1 (VIA4), Δatp23 (CW3) or Δyme1Δatp23 (CW79) cells and the stability of newly imported Ups1 upon further incubation at 37°C was assessed by SDS–PAGE and autoradiography. Quantification of Ups1 accumulation in mitochondria is shown in the lower panel. Newly imported Ups1 was set to 100%. Data represent ±s.d. of four independent experiments. *P<0.05, **P<0.005, ***P<0.0005. (D) Steady-state levels of Ups1 and Ups2 in mitochondria lacking Yme1 and Atp23. Mitochondria were isolated from WT (CG), Δatp23 (CW3), Δyme1 (VIA4), Δatp23Δyme1 (CW79), Δyme1Δmdm35 (CW414) and Δmdm35 (CG323) cells and analysed by SDS–PAGE and immunoblotting using Yme1-, Tom40- and affinity-purified Mdm35-, Ups1- and Ups2-specific antibodies. (E) Quantitative assembly of Ups1 and Ups2 with Mdm35 in Δyme1 mitochondria. Sucrose-gradient purified mitochondria isolated from Δyme1 cells were solubilized in 1% (w/v) dodecylmaltoside (DDM) at a protein concentration of 5 mg/ml. After a clarifying spin, extracts were subjected to sizing chromatography using a Superose 12 column (GE Healthcare), which had been equilibrated with 0.1% (w/v) DDM, 25 mM Tris/HCl pH 7.4, 150 mM NaCl, 10% (v/v) glycerol. Eluate fractions were TCA-precipitated and analysed by gradient Tris/Tricine gradient SDS–PAGE and immunoblotting using Ups1-, Ups2- and Mdm35-specific antisera. (a) alcoholdehydrogenase (150 kDa), (b) ovalbumine (44.3 kDa), (c) cytochrome c (12.4 kDa). The asterisk (*) refers to a band unspecifically cross-reacting with the respective antiserum (D, E).