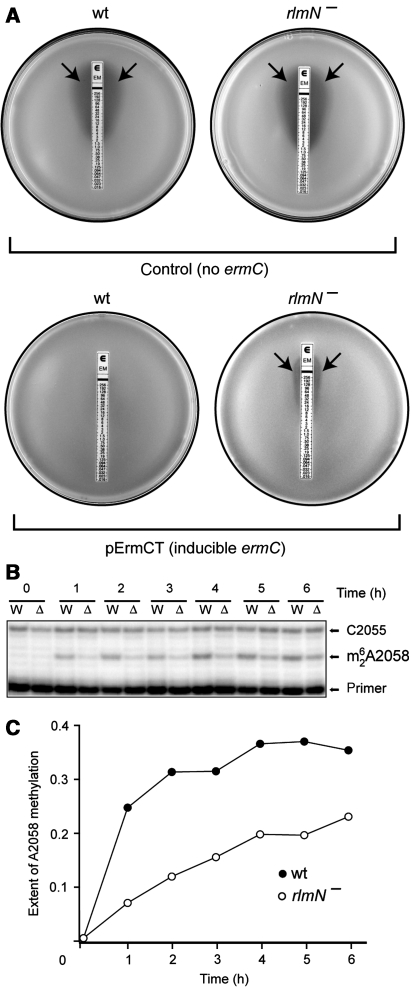
Figure 2.
The effects of indigenous posttranscriptional modification of A2503 upon ermC induction. (A) The E-test analysis of erythromycin resistance of the E. coli cells that either do not carry the ermC gene (control) or carry a plasmid pErmCT with inducible ermC. The rlmN+ cells (wt) or rlmN− cells (lacking posttranscriptional modification of A2503) were plated onto agar plates and overlaid with an E-strip containing a gradient of erythromycin concentration. Inhibition of cell growth is manifested as a clear zone around the strip (arrows). (B) Primer-extension analysis of the induction of 23S rRNA modification by ErmC upon exposure of cells to erythromycin. Exponential wild-type (W) or rlmN− (Δ) cells carrying inducible ermC were induced with 32 μg/ml erythromycin, and the extent of ErmC-catalysed A2058 dimethylation in 23S rRNA at specified time points was analysed by primer extension. In the presence of ddGTP terminator, reverse transcriptase stops at the dimethylated A2058 but advances to C2055 when A2058 remains unmodified. (C) Quantification of the intensities of the primer-extension bands on the gel shown in (B), representing the fraction of the modified 23S rRNA.