Figure 3.
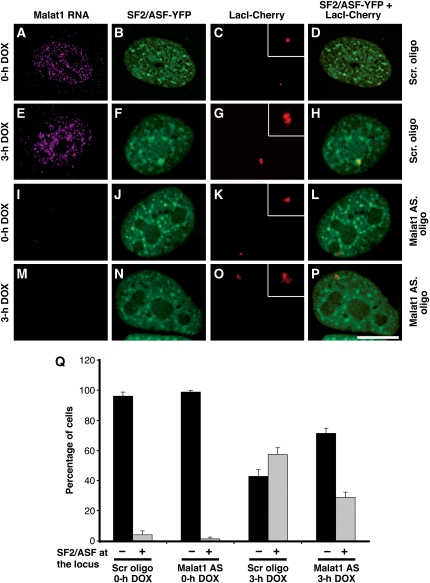
Malat1 ncRNA facilitates the recruitment of SF2/ASF to an active transcription site. (A–D) RNA-FISH to U2OS 2-6-3 cells (Janicki et al, 2004) stably expressing LacI-mCherry and rtTa transactivator (Tet-ON) shows a punctate nuclear localization of Malat1 ncRNA. Note the more homogenous nuclear staining pattern of Malat1 ncRNA in U2OS cells (A, E). In several of cancer cell lines examined, a population of cells (20–25%) showed a nuclear speckle pattern of Malat1 ncRNA unlike primary diploid cell lines and tissues where >70% of the cells exhibited a speckled distribution of Malat1 ncRNA. Cells treated with a scrambled oligonucleotide (A–D) or Malat1-specific oligonucleotide (I–L) in the absence of doxycycline (0 h DOX) do not show SF2/ASF (B, J) at the transcriptionally inactive reporter gene locus (C, K, D, L). (E–H) Upon addition of doxycycline (3 h DOX), the transcriptionally active locus (G) showed enrichment of SF2/ASF (F, H). (I–P) Cells treated with Malat1-specific antisense oligonucleotides showed complete depletion of Malat1 ncRNA (I, M). In the absence of Malat1 ncRNA, upon addition of doxycycline (3 h DOX), a significantly reduced level of SF2/ASF (N, P) was associated with the transcriptionally active locus (O). The inset in figures (C, G, K, O) represents the magnified reporter locus. Scale bar, 5 μm. (Q) The histogram shows the percentage of cells that exhibit recruitment of SF2/ASF to the transcriptionally inactive or active reporter locus in the presence or absence of Malat1 ncRNA. The data represents mean and s.d. values of three independent experiments per data point (n=25 cells/experiment).