Abstract
The nonnucleoside reverse transcriptase inhibitors (NNRTIs) are key components of highly active antiretroviral therapy (HAART) for the treatment of human immunodeficiency virus type 1 (HIV-1). A major problem with the first approved NNRTIs was the emergence of mutations in the HIV-1 reverse transcriptase (RT), in particular K103N and Y181C, which led to resistance to the entire class. We adopted an iterative strategy to synthesize and test small molecule inhibitors from a chemical series of pyrazoles against wild-type (wt) RT and the most prevalent NNRTI-resistant mutants. The emerging candidate, lersivirine (UK-453,061), binds the RT enzyme in a novel way (resulting in a unique resistance profile), inhibits over 60% of viruses bearing key RT mutations, with 50% effective concentrations (EC50s) within 10-fold of those for wt viruses, and has excellent selectivity against a range of human targets. Altogether lersivirine is a highly potent and selective NNRTI, with excellent efficacy against NNRTI-resistant viruses.
Since it was first reported in 1981, AIDS has rapidly become a pandemic fatal disease; in 2007, UNAIDS estimated that between 30 and 36 million people globally were infected with human immunodeficiency virus type 1 (HIV-1) and that every day more than 6,800 people become infected with HIV-1 while more than 5,700 people die from AIDS (45). To date, no cure exists to eliminate HIV.
The treatment of care known as highly active antiretroviral therapy (HAART) consists of a combination of at least three selected antiretroviral drugs (taken from six different drug classes) that inhibit the HIV replication cycle at the point of entry (chemokine antagonists, fusion inhibitors), reverse transcription (nucleoside/nucleotide and nonnucleoside reverse transcriptase inhibitors), integration (integrase inhibitors), or viral production (protease inhibitors [PIs]).
The first drugs to be approved in the nonnucleoside reverse transcriptase inhibitor (NNRTI) class were delavirdine (18, 40), nevirapine (NVP) (22, 31), and efavirenz (8, 29). All three drugs are efficacious at inhibiting HIV replication, but their use and compliance are limited by serious side effects, frequently leading to the rapid generation of resistance through single point mutations which can confer class resistance (29, 41). When observed in the clinical setting, such mutations preclude the sequential use of NNRTIs in HAART regimens (3).
Second-generation NNRTIs, which are either licensed or in clinical development, retain activity against clinically significant drug-resistant viruses (2, 27). Most notably, clinical trials with etravirine (TMC125) demonstrate that such second-generation NNRTIs can contribute to virologic suppression in NNRTI-experienced patients and can be useful additions to the armamentarium of drugs for the treating physicians. However, the use of etravirine in first-line patients is likely to be hampered by the potential for drug-drug interactions and a relatively high rate of adverse effects (rash and nausea in particular), as well as severe skin and hypersensitivity reactions, including cases of hepatic failure (44). The efficacy of etravirine in treatment-naïve patients remains to be established.
Efavirenz remains the current NNRTI of choice, with efavirenz-based therapy or boosted PI-based therapy in combination with nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs) currently recommended as first-line regimens for the treatment of HIV infection. Well-tolerated, conveniently administered NNRTIs with activity against drug-resistant virus could be equally useful in treating previously untreated patients as well as those with a history of NNRTI failure.
In this report, we present the preclinical profile of a novel and potent NNRTI, designated lersivirine (UK-453,061), which resulted from lead optimization of a substituted pyrazole. Lersivirine has a novel mode of binding to the enzyme, resulting in largely nonoverlapping resistance with existing agents from the class. It is exquisitely selective against a range of human targets, has broad-spectrum activity against primary and lab-derived NNRTI-sensitive or NNRTI-resistant strains, and appears to be additive or synergistic when used in combination with other agents. Emergence of resistance to lersivirine requires multiple mutations, and the resistant viruses are mostly sensitive to first-generation NNRTI drugs. These antiviral results, along with other preclinical data available to date, indicate that lersivirine should be a convenient and effective therapy that is well tolerated by patients.
MATERIALS AND METHODS
Compounds.
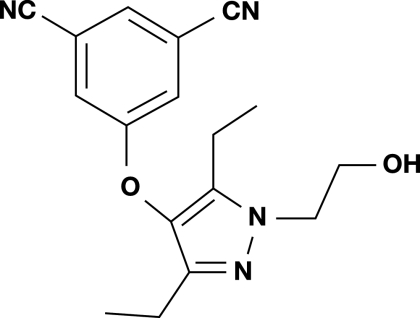
Lersivirine [5-([3,5-diethyl-1-(2-hydroxyethyl)-1H-pyrazol-4-yl]oxy)isophthalonitrile] (structure shown in Fig. 1) was synthesized at Pfizer Global Research and Development Laboratories. Enfuvirtide was synthesized by NeoMPS (Strasbourg, France). All other anti-HIV compounds were synthesized at Pfizer Global Research and Development Laboratories. For combination studies, abacavir, emtricitabine, tenofovir, and ritonavir were obtained from Moravek Biochemicals, Brea, CA; didanosine (ddl) was purchased from Sigma, Dorset, United Kingdom; and lamivudine (3TC) was obtained from APIN Chem Ltd., Oxford, United Kingdom. For antiviral assays, inhibitors were dissolved in dimethyl sulfoxide (DMSO).
FIG. 1.
Structure of lersivirine.
Cells and viruses.
All cell lines and viruses, except where stated, were obtained from the Centralized Facility for AIDS Reagents, National Institute for Biological Standards and Control (NIBSC, Potters Bar, Hertfordshire, United Kingdom). The indicator cell line HeLaP4, expressing human recombinant CD4, CCR5, and CXCR4, was kindly provided by Ned Landau (Aaron Diamond AIDS Research Center, New York, NY). HeLaP4 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated fetal calf serum (FCS), 2 mM l-glutamine, 1 U/ml penicillin, and 0.1 mg/ml streptomycin. HEK293 (human embryonic kidney) cells were maintained in DMEM containing 10% heat-inactivated FCS, 2 mM l-glutamine, 1% nonessential amino acids (NEAAs), 1 mM sodium pyruvate, 1 U/ml penicillin, and 0.1 mg/ml streptomycin. Puromycin, at a final concentration of 0.5 μg/ml, was included in all HeLaP4 cultures. MT-2 (human T-cell leukemia) and SupT1 (non-Hodgkin's T-cell lymphoma subclone) cells were cultured in RPMI 1640 medium containing 10% heat-inactivated FCS, 2 mM l-glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin. Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Paque gradient separation (Amersham Pharmacia Biotech, Piscataway, NJ) from pooled buffy coats (North London and South London Blood Transfusion Services, United Kingdom) from three or four HIV-1- and hepatitis B virus-seronegative donors per batch. PBMCs were stimulated with phytohemagglutinin (PHA; Murex, Abbott Laboratories) at 1.5 μg/ml for 3 days in RPMI 1640 medium containing heat-inactivated 10% FCS, 2 mM l-glutamine, 1 U/ml penicillin, and 0.1 mg/ml streptomycin. All cells were cultured at 37°C in a humidified incubator with a 5% CO2 atmosphere.
NL4-3 infectious virus was generated following transfection of the plasmid pNL4-3 (National Institute of Health AIDS Research and Reference Reagent Program; https://www.aidsreagent.org), which contains a full-length HIV-1 genome constructed from two North American isolates (5′ sequence derived from NY5 and 3′ sequence derived from LAV) (1) into HEK293 cells. Similarly, NL4-3 mutant viruses, modified to contain NNRTI resistance-associated mutations, were generated by transfection of the mutated plasmid. NL4-3 virus stocks were expanded by limited passaging in SupT1 cells. Stocks of the HIV-1 laboratory-adapted strain Ba-L and primary isolates 92BR017 (clade B), 98IN022 (clade C), 93BR029 (clade F), and RU570 (clade G) were prepared by limited passaging in PHA-stimulated PBMCs.
Site-directed mutants.
DNA encoding the histidine-tagged reverse transcriptase (RT) (p66) and protease genes (HIV-1, BH10 strain) were cloned into the expression vector pT5m and kindly provided by Stephen Hughes (19). Specific amino acid replacements were made using the QuikChange site-directed mutagenesis kit (Stratagene). Following mutagenesis, the entire mutant RT coding sequence was verified by sequencing the corresponding plasmids. RT plasmids containing mutations at the following locations were constructed: L100I, K101E, V106A, V108I, E138K, Y181C, Y181I, M184V, F227L, F227C, E233V, L234I, P236L, V106A/F227L, Y181C/Y188C, Y181I/Y188L, and V106A/Y181C. The mutated RT genes were used to construct recombinant NL4-3 mutant viruses by replacing the RT gene in pNL4-3 with the corresponding gene from the mutant pT5m-RT clone. Plasmids were sequenced to verify mutations (Lark Technologies, Essex, United Kingdom).
RT expression and purification.
For expression, either wild-type (wt) or mutant RT plasmids were transfected in supercompetent BL21(DE3) cells (Stratagene). After induction with isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM final), wt protein and each mutant protein were purified from bacterial lysates using nickel chelate affinity chromatography (Hi-Trap chelating column) followed by Mono Q chromatography. Fractions containing p66/p51 heterodimeric RT were pooled, concentrated using Centriprep 30, and stored at −80°C.
RT enzyme assays.
The activities of lersivirine and efavirenz against wt and mutant HIV-1 RT (BH10 strain) were determined using a primer extension assay incorporating a DNA/RNA primer/template as described previously (25). The 5′ biotinylated primer DNA is a 16mer oligo(dT), which is annealed to a poly(rA) template of approximately 300 bases in length. Incorporation of [3H]TTP (dTTP) by reverse transcription results in extension of the primer. During the reaction the primer/template is bound to a streptavidin-coated flashplate (NEN). The incorporated tritiated nucleotides can then stimulate the scintillant to produce a signal that is measured using a scintillation counter. Compounds that inhibit the RNA-dependent DNA polymerase activity of RT produce a reduced signal. A dose response curve can then be used to calculate the 50% inhibitory concentration (IC50).
ITC.
Recombinant HIV-1 RT was prepared to suitable concentrations for isothermal titration calorimetry (ITC) (typically 13.2 to 13.9 μM) using Microcon-10 microconcentrators (Amicon). ITC experiments were performed twice at 25°C in 5% DMSO using a MicroCal VP-ITC, with 215 μM lersivirine titrated into HIV-1 RT over 25 injections of 10 μl, made every 230 s, into a fixed reaction cell volume of 1.4272 ml.
X-ray crystallography.
Heterodimeric wt and K103N HIV-1 RT were expressed and purified as described previously (28) and crystallized in the presence of lersivirine. The crystals belong to space group C2221 (unit cell dimensions: a ≈ 117 Å, b ≈ 154 Å, and c ≈ 155 Å) and are isomorphous with those reported previously (24). X-ray diffraction data, to 2.8 Å and 3.2 Å resolution, were collected at 100 K with an ADSC Quantum 4 CCD detector at Station 14.2 of SRS, Daresbury, United Kingdom, and “in house” using a Rigaku FRD generator and Saturn92 detector. Data processing and scaling were carried out using the CCP4 suite of programs (5). The structures were determined by molecular replacement and difference. Fourier methods using the coordinate set 1 fkp (36) as the starting model. Interactive graphical model building was carried out with Quanta (Accelrys Inc., San Diego, CA) and Coot (14), with iterative rounds of refinement carried out using CNX (Accelrys Inc., San Diego, CA) and Buster (39).
In both structures the ligand was well defined by the initial electron density maps. In refinement, the option of applying additional restraints to a previous higher-resolution target structure (available in Buster 1.7.2) was used, and only group B factors were refined for the lower-resolution structure. Data and refinement statistics are listed in Table S1 in the supplemental material.
Human DNA polymerase β assay.
The activity of lersivirine against human DNA polymerase was determined using a primer extension assay. A 5′ biotinylated 16mer oligo(dT) primer DNA was annealed to a poly(dA) template and was extended during incubation with human DNA polymerase. Incorporation of [3H]TTP was detected by scintillation.
Drug susceptibility assays.
The antiviral activity of lersivirine against the HIV-1 strain NL4-3 wt and a panel of NL4-3 recombinant viruses with NNRTI resistance-associated mutations was determined using the indicator cell line, HeLaP4. The adherent HeLaP4 cells were detached with cell dissociation solution (CDS; Sigma), resuspended in culture medium, and exposed to virus. Infected cultures were added to triplicate wells of a 96-well plate at 4.5 × 103 cells/well containing serial dilutions of test compound in culture medium. After a 5-day culture, virus replication was quantified by measuring levels of HIV-1 Tat-induced galactosidase (FluorAce β-galactosidase reporter assay; Bio-Rad).
Drug susceptibility testing of the laboratory-adapted strain Ba-L and primary isolates was performed in PHA-stimulated PBMCs. Cells were infected for 1 h at 37°C using a predetermined volume of virus calculated to give comparable multiplicities of infection (MOI) (0.01 to 0.1) or equivalent amounts of RT activity per infection. Infected cells were washed and resuspended in medium containing 10 ng/ml human recombinant interleukin-2 (IL-2) (R&D Systems) and added to duplicate wells of 24-well plates (3.6 × 105 cells/well) or triplicate wells of 96-well plates (3.6 × 104 cells/well) containing serial dilutions of test compound in culture medium. After 5 to 7 days of incubation, virus replication was quantified by measuring supernatant p24 levels (HIV monoclonal antibody [MAb] p24 enzyme-linked immunosorbent assay [ELISA]; Murex, Abbott Diagnostics) or RT activity (Quant-T-RT assay; GE Healthcare). Any compound-specific cytotoxicity was assessed in parallel (see “Cytotoxicity testing” below).
For all drug susceptibility assays, the percent virus inhibition at each compound concentration was calculated and sigmoidal dose-response curves were fitted using a proprietary curve-fitting program to determine the anti-HIV-1 50% and 90% effective concentrations (EC50 and EC90, respectively).
MOI assays.
Antiviral drug susceptibility assays were performed in MT-2 cells to assess the in vitro effect of virus input on antiviral potency. Briefly, cells were infected with a predetermined volume of virus calculated to an MOI of 0.005, 0.05, or 0.5 50% tissue culture infective dose (TCID50) per cell. Infected cells were added to duplicate wells of 96-well plates (1.8 × 104 cells/well) containing serial dilutions of test compound in culture medium. After 6 days of incubation, virus replication was measured indirectly by assessing cell viability (Cell Titer-Glo; Promega). The NNRTIs etravirine and efavirenz, the PI indinavir, and the investigative maturation inhibitor PA-457 were included as controls in the assay. The percent inhibition of virus-induced cell death for each concentration at each MOI was used to determine the anti-HIV-1 EC50 and EC90.
Drug combination studies.
The in vitro antiviral effect of lersivirine in combination with representatives of the NRTIs (abacavir, didanosine, emtricitabine, lamivudine, tenofovir, and zidovudine), the PIs (atazanavir, lopinavir, and ritonavir), the entry inhibitor (enfuvirtide), or the integrase inhibitors (raltegravir and elvitegravir) was evaluated in HIV-1 NL4-3 wt acutely infected MT-2 cells (MOI, 0.05 to 0.1 TCID50 per cell). Infected cells (8 × 103 cells/well) were plated into a 384-well assay plate containing serial dilutions of lersivirine and the test compound prepared in 0.05% Pluronic acid (Sigma). Virus growth was determined indirectly after a 5-day culture by measuring the metabolic activity of viable cells (Cell Titer-Glo luminescent cell viability assay kit; Promega). Data were analyzed using the MacSynergy II three-dimensional model of Prichard and Shipman (34). Combinations of test compound titrated against itself were assayed in parallel; lersivirine data were reported only when the “sham” combinations indicated additive interactions.
Phenotypic resistance profiling.
Drug susceptibility was determined using cell-based pseudovirus assays at Virco (Antivirogram assay; Mechelen, Belgium) and Monogram Biosciences Inc. (PhenoSense HIV assay; South San Francisco, CA). In both assays, the HIV-1 RT gene is amplified from virus samples by PCR analysis and the resultant amplicons are inserted into HIV-1-derived expression vectors lacking the RT gene. Through a process of gene transfer (Nucleofectin; Virco) or cotransfection with an expression vector encoding the Env proteins (Monogram Biosciences Inc.), infectious virus particles are produced. The ability of the pseudovirus to infect target cells in the presence or absence of various concentrations of inhibitors is measured. The Virco Antivirogram assay was used to determine susceptibility against a panel of 191 HIV-1 isolates derived from the plasma of treatment-naïve and antiretroviral-treatment-experienced individuals chosen to identify a comprehensive phenotypic resistance profile for lersivirine. Three different virus panels were investigated at Monogram Biosciences using the PhenoSense HIV assay: 100 HIV-1 isolates derived from NNRTI treatment-experienced patients failing on a PI regimen, 62 HIV-1 isolates derived from NNRTI treatment-naïve patients with evidence of genotypic (transmitted) resistance to efavirenz, 45 HIV-1 isolates derived from NNRTI treatment-naïve patients representing different viral clades (A, A1, B, C, D, F1, and G), circulating recombinant forms (CRF) (AE, AG, and BF), and recombinant forms (C/H). Twenty-five of the viruses were clade C, 5 were clade B, 5 were CRF BF, and the remaining were A, A1, AE, C/H, D, F1, and G.
The distribution of NNRTI mutations in 353 clinically derived recombinant HIV isolates (191 isolates from Virco combined with the 100 and 62 isolates from Monogram Biosciences) is shown in Table 1. It should be noted that the proportion of each mutant in the panel of viruses tested may not reflect the incidence of these mutants in the HIV-1 drug-experienced patient population. Mutations at the following locations in RT have been previously described as conferring resistance to NNRTIs: A98G, L100I, K101E, K103N, V106A, V108I, V179D/E, Y181C/I, Y188L/C/H, G190A/S/E/Q, P225H, F227L, M230L, L234I, P236L, and Y318F (23, 30, 42). In total, 488 NNRTI mutations at the codons listed above were detected in 228 of the 353 isolates (Table 1). In some virus isolates, a particular codon contained a mixture of NNRTI mutant and wt sequences, indicating the presence of multiple viral quasi-species in the sample. For the purposes of analysis, these positions were scored as containing an NNRTI mutation. Linkages between mutations in viruses containing >1 NNRTI mutation are not represented.
TABLE 1.
Prevalence of NNRTI-specific mutations in recombinant HIV-1 isolatesa
| Mutation | No. of viruses containing mutationb | % of total mutationsc | No. of mutation mixturesd |
|---|---|---|---|
| A98G | 24 | 5 | 7 |
| L100I | 24 | 5 | 4 |
| K101E | 23 | 5 | 6 |
| K103N | 166 | 34 | 18 |
| V106A | 12 | 2 | 3 |
| V108I | 23 | 5 | 7 |
| V179D | 5 | 1 | 2 |
| Y181C/I | 71 | 15 | 11 |
| Y188L | 17 | 3 | 3 |
| Y188C | 8 | 2 | 3 |
| G190A/S | 59 | 12 | 12 |
| G190E/Q | 8 | 2 | 1 |
| P225H | 16 | 3 | 6 |
| F227L | 10 | 2 | 1 |
| M230L | 11 | 2 | 3 |
| L234I | 1 | 0 | 0 |
| P236L | 2 | 0 | 1 |
| Y318Fe | 8 | 2 | 1 |
| Total | 488 | 100 | 89 |
Mutations present in the population at a low frequency (e.g., Y188H, K101P, etc.) were not included in this analysis.
Singly or in combination with other NNRTI-specific mutations.
n = 488.
Mixture of mutant and wt virus detected.
Mutation Y318F was not investigated at Monogram Biosciences.
BCO.
The biological cutoff (BCO) for virus susceptibility has been calculated using the same method as described previously (48). Applying this method to the 63 wt clinical isolates tested here, the calculated BCOs are 3.1 for lersivirine, 1.75 for efavirenz, 6.63 for delavirdine, and 5.86 for nevirapine. The BCO values were subsequently used to establish sensitivity.
In vitro resistance selection.
For inhibitor escalation studies, SupT1 cells were initially infected with HIV-1 NL4-3 wt (MOI, 7.4 TCID50 per cell) for 1 h at 37°C and the virus-infected cells were passaged weekly in SupT1 cells in the presence of increasing concentrations of lersivirine from a starting concentration of 5 nM (IC50 in this assay system) in 12-well plates (6 × 105 cells/well). Throughout the passaging period, ongoing virus replication was monitored weekly by observing cytopathic effects (syncytia). Every 7 days, virus supernatant was passaged onto fresh cells at the same density with fresh lersivirine-containing medium. The concentration of lersivirine was increased by 2-fold and 5-fold that of the previous concentration upon subsequent passage whenever evidence of viral replication was observed. Virus stocks for phenotypic characterization were produced in SupT1 cells in the absence of compound and were tested for their sensitivity to lersivirine in an antiviral assay using HeLaP4 cells as previously described. In addition, virus supernatants (harvested during passaging) were sent to Monogram Biosciences for testing in their PhenoSense and GeneSeq assays. Drug-free passages were set up in parallel to control for changes in sensitivity to lersivirine upon prolonged culturing of virus. In addition, virus passaging in the presence of efavirenz was set up in parallel.
For fixed-dose studies, SupT1 cells were initially infected with HIV-1 NL4-3 wt or the RT site-directed K103N and Y181C mutants (MOI, 0.1 TCID50 per cell) for 1 h at 37°C and the virus-infected cells cultured in the presence of fixed concentrations of lersivirine representing a range of EC50 multiples (10×, 20×, and 100×; EC50, 9.52 nM in this assay system) in 24-well plates (3.6 × 105 cells/well). Cultures were fed fresh uninfected cells and fresh compound, maintaining the same final compound concentration, every 7 days. Throughout the passaging period (21 days), virus replication was quantified by observing syncytia. Breakthrough virus was harvested when >25% of the culture showed evidence of syncytia formation. The virus supernatant was sent to Monogram Biosciences for testing in their PhenoSense and GeneSeq assays. Drug-free passages were set up in parallel to control for changes in sensitivity to lersivirine upon prolonged culture of virus. In addition, virus was passaged in the presence of efavirenz and etravirine.
Cytotoxicity testing.
The potential toxic activity of lersivirine in HeLaP4 cells and peripheral blood lymphocytes (PBL) was determined using the cell titer 96 aqueous nonradioactive cell proliferation assay (Promega). In these assays, HeLaP4 cells or PBL were cultivated for 5 days under conditions identical to those used in their respective antiviral assays, except that no virus was added to the cells. Assays were performed according to the manufacturer's instructions using a range of concentrations of lersivirine (with a maximum concentration of 200 μM).
Activity of lersivirine against potential human target proteins in selectivity screening assays.
The specificity of lersivirine was tested against various receptors and enzymes using Cerep's BioPrint service (see the Cerep website for more information). The list of targets against which lersivirine was profiled is presented in the supplemental material.
Protein Data Bank accession numbers.
Coordinates and reflection files were deposited in the Protein Data Bank under accession numbers 2 won and 2 wom.
RESULTS
Lersivirine is a potent and selective nonnucleoside reverse transcription inhibitor.
Lersivirine and efavirenz were initially tested (n = 20) for in vitro inhibitory activity against purified wt RT using a primer extension assay, and both were found to be potent inhibitors of the RT activity of HIV-1 wt RT, with geometric mean IC50s of 118 nM (95% confidence interval [CI], 100 to 139 nM) and 7.2 nM (95% CI, 4.4 to 11.9 nM) for lersivirine and efavirenz, respectively.
The binding affinity of lersivirine to RT was characterized using isothermal calorimetry (ITC) (Fig. 2). Two independent ITC experiments were conducted, demonstrating that lersivirine binds to recombinant HIV-1 RT at 25°C with a mean geometric dissociation constant (KD) of 624 nM (standard error, 42.5 nM). The derived stoichiometry of the interaction is consistent with a 1:1 binding mechanism. It is interesting to note that the binding of lersivirine is enthalpy driven (ΔH [change in enthalpy] = −2.305 × 104 cal/mol ± 3.01 × 103; ΔS [change in entropy] = −48.9 cal/K/mol ± 9.9). This indicates that upon binding of lersivirine to the viral RT there is a decrease in entropy of the system, possibly due to a decrease in the mobility of a portion of the protein.
FIG. 2.
Typical example of a titration of lersivirine into recombinant wt HIV-1 RT at 25°C in 5% dimethyl sulfoxide. In this example, KD was 668 nM ± 12%, ΔH was −2E4 ± 528.9, and ΔS was −38.97.
Consistent with the findings described above, the mechanism of inhibition of the RT enzyme by lersivirine was confirmed in kinetic experiments as “mixed noncompetitive” (as for other NNRTIs), with a geometric mean Ki value of 117 nM and a geometric mean Ki value of 340 nM (data not shown). Finally, the effect of increasing viral MOI on the antiviral potency (EC50) of lersivirine following infection of MT-2 cells with wt NL4-3 is summarized in Table 2. We showed that lersivirine is able to inhibit HIV-1 virus replication, with an EC50 ranging from 5 nM to 35 nM against an MOI ranging from 0.005 to 0.5. A shift of <10-fold in potency across a 100-fold virus input is similar to that observed for two other NNRTIs, efavirenz and etravirine, tested in parallel, and consistent with its mode of action (MOA) being in the early stages of viral replication. In contrast, indinavir (a PI) and PA-457 (a maturation inhibitor) demonstrated shifts of >130 and >101, respectively, in the same assays.
TABLE 2.
Impact of viral input on compound EC50 across a 100-fold range of MOI
| Compound (n) | MOI | EC50 (nM) | Lower 95% CI (nM) | Upper 95% CI (nM) | EC50 fold change over MOI range |
|---|---|---|---|---|---|
| Lersivirine (5) | 0.005 | 5.04 | 1.9 | 13.0 | 6.9 |
| 0.05 | 13.1 | 5.99 | 28.6 | ||
| 0.5 | 34.9 | 18.1 | 67.3 | ||
| Efavirenz (5) | 0.005 | 0.515 | 0.228 | 1.17 | 4.3 |
| 0.05 | 0.908 | 0.437 | 1.89 | ||
| 0.5 | 2.20 | 1.42 | 3.41 | ||
| Etravirine (3) | 0.005 | 1.56 | 0.343 | 7.12 | 2.9 |
| 0.05 | 2.58 | 0.780 | 8.55 | ||
| 0.5 | 4.56 | 2.24 | 9.30 | ||
| Indinavir (3) | 0.005 | 4.87 | 1.35 | 17.6 | >130 |
| 0.05 | 12.7 | 3.77 | 42.6 | ||
| 0.5 | >500 | NAa | NA | ||
| PA-457 (5) | 0.005 | 39.6 | 19.5 | 80.6 | >101 |
| 0.05 | 119 | 74.2 | 191 | ||
| 0.5 | >4,000 | NA | NA |
NA, not available.
To confirm the selectivity of lersivirine for HIV-1 RT, we evaluated the activity of lersivirine against human DNA polymerase beta. Lersivirine is a very weak inhibitor of human DNA polymerase beta, with an extrapolated geometric mean IC50 of approximately 20 mM resulting in a predicted selectivity index (defined as the IC50 against DNA polymerase beta divided by the IC50 against RT) of 166,000. An additional 28 human receptors and enzymes were also profiled for susceptibility to lersivirine, with no human target proteins being significantly modulated at concentrations of up to 10 μM (data not shown), with the exception of the human phosphodiesterase IV, which displayed an IC50 of 13 μM. Finally, the cytotoxicity of lersivirine for exponentially growing HeLaP4 cells as well as primary blood cells was determined using a cell titer 96 aqueous nonradioactive cell proliferation assay. No cell cytotoxicity was observed at the highest concentration of lersivirine tested in HeLaP4 cells (200 μM), while in PBMCs lersivirine had a median 30% cytotoxicity (CC30) of greater than 96.9 μM (lower quartile = 82.4 μM; upper quartile > 200 μM). The ratio between the CC30 and the antiviral EC90 is greater than 7,000 (>7,092 in HeLaP4 cells and equal to 9,818 in PBL).
Lersivirine binds HIV-1 RT in a novel binding mode and retains activity against commonly encountered mutations in the RT enzyme.
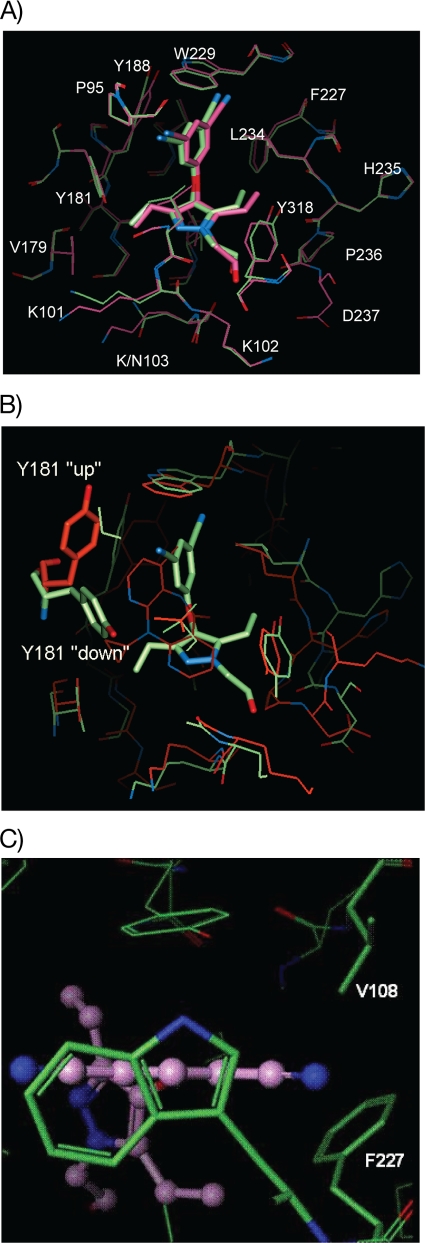
The binding mode of lersivirine to the recombinant wt HIV-1 RT protein (strain HXB2) was further investigated using protein X-ray crystallography. The X-ray structure confirmed that lersivirine binds noncovalently to the allosteric, nonnucleoside binding site of recombinant wt HIV-1 RT, as illustrated in Fig. 3A.
FIG. 3.
Crystal structure of lersivirine bound to HIV-1 RT. (A) Lersivirine bound at the nonnucleoside binding site of recombinant HIV-1 RT. pink, wt; green, K103N mutant. (B) Overlay of NNRTI binding pocket for lersivirine (green) and nevirapine (red), showing the change in conformation of Y181. (C) wt structure viewed looking down on W229 in the plane of the disubstituted phenol ring of lersi- virine. The residues F227 and V108, the positions of resistance mutations, are highlighted. These residues make apolar van der Waals packing interactions with the compound.
Detailed analysis of this binding indicates that lersivirine forms interactions with residues L100, V106, Y181, Y188, F227, W229, Y318, L234, and P236 of the p66 subunit of RT. Interestingly, Y181 is rotated approximately 100° around χ1 with respect to the conformation found in complexes with nevirapine or efavirenz (Fig. 3B). This Y181 rotation in lersivirine-bound RT protein is similar to the rotation found in the apostructure. V108 makes extensive van der Waals interactions with the dicyano-substituted phenol moiety of lersivirine. Mutations to larger residues at the 108 position are predicted to perturb the observed binding mode (Fig. 3C). The binding mode of lersivirine to the key K103N mutant of HIV-1 RT was then investigated using the same technique. As depicted in Fig. 3A, the binding of lersivirine to the mutant enzyme is similar to the binding mode of lersivirine to the wt enzyme, consistent with lersivirine retaining activity against this key NNRTI-resistant mutation. Indeed, when tested in the RT elongation assay, lersivirine inhibited K103N RT with an IC50 of 215 nm (95% CI, 150 to 309%), which is within 2-fold of that observed for the wt enzyme. In contrast, efavirenz inhibited K103N RT with an IC50 of 414 nm (95% CI, 268 to 640%), a marked reduction of approximately 60-fold compared with that of wt RT and in agreement with what has been reported previously (50).
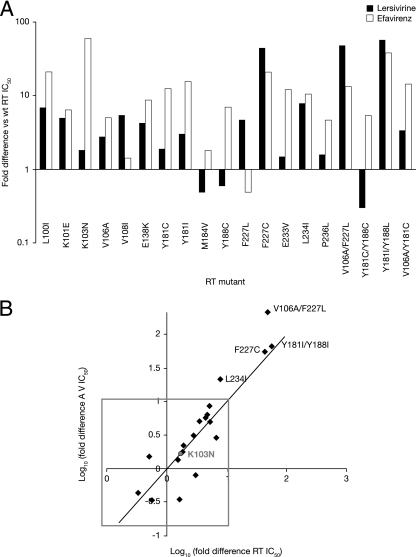
These experiments were then extended to include a panel of 19 single and double point mutant RTs (Fig. 4A). The mutations were chosen to represent those most frequently found in samples from NNRTI-experienced patients (32, 42, 47). Lersivirine was shown to inhibit 14 out of 15 single point mutant RTs (L100I, K101E, K103N, V106A, V108I, E138K, Y181C, Y181I, M184V, Y188C, F227L, E233V, L234I, and P236L) with IC50s within 10-fold of those measured for the wt enzyme, compared with only 8 out of 15 for efavirenz. The activity of both efavirenz and lersivirine was significantly reduced against F227C (20.2-fold and 44.3-fold increase in IC50, respectively). In addition to the activity against the mutant described above, the activity of efavirenz was also significantly reduced against L100I, Y181C, Y181I, E233V, and L234I RT enzymes, consistent with previous reports (10, 12).
FIG. 4.
Activity of lersivirine against a panel of mutant RT enzymes and viruses. (A) Lersivirine and efavirenz were tested against a panel of mutant RT enzymes, and their respective fold difference versus wt RT was plotted. White bars, efavirenz; black bars, lersivirine. (B) xy plot of the log of the fold difference of the activity of lersivirine against mutant RTs versus the log of the activity of lersivirine against the corresponding mutant NL4-3 viruses.
Of the four double mutants tested, the Y181C/Y188C mutant was sensitive (a <10-fold increase in IC50 compared to that for the wt) to both efavirenz and lersivirine, while lersivirine alone was active against the V106A/Y181C double mutant RTs. Both lersivirine and efavirenz had reduced activity against the double mutant V106A/F227L (47.9- and 13.1-fold drop in potency, respectively) and Y181I/Y188L (56.5- and 37.4-fold drop in potency, respectively) RTs.
The same mutations were then introduced into the infectious recombinant NL-4.3 virus background, and their susceptibilities to lersivirine and efavirenz were determined on an indicator HeLaP4 cell line, measuring levels of HIV-1 Tat-induced β-galactosidase 5 days following infection (Fig. 4B). Lersivirine inhibited replication of the wt virus, with a geometric mean EC50 of 3.96 nM, while the EC50 for the K103N mutant virus was 7.05 nM. Fifteen of the 19 mutated viruses investigated (L100I, K101E, K103N, V106A, V108I, E138K, Y181C, Y181I, M184V, Y188C, F227L, E233V, P236L, V106A/Y181C, and Y181C/Y188C) were sensitive to inhibition by lersivirine, with an EC50 within 10-fold of that for the wt virus, including the amino acid substitution M184V, which commonly arises in response to treatment with NRTI. Reduced susceptibility to lersivirine was seen with the substitutions L234I (∼20-fold), F227C (∼55-fold), Y181I/Y188L (∼65-fold), and V106A/F227L (>200-fold), which is consistent with the effect of the same substitutions on the activity of lersivirine against the RT enzyme (Fig. 4B).
Lersivirine inhibits clinically relevant viral strains.
To further extend the antiviral characterization of lersivirine, its activity was evaluated against three panels of viruses containing clinically relevant mutations. In these panels, the compound was assessed against 353 clinically derived recombinant viruses using either Virco's Antivirogram assay (191 clinical isolates) or Monogram Bioscience's PhenoSense HIV assay (162 clinical isolates). Sensitivity was assessed using the BCO values calculated for each compound, and the results are summarized in Tables 3 and 4. Of the 353 viruses tested, 290 contained one or more NNRTI-specific mutation (summarized in Table 1). Out of 63 viruses with no NNRTI mutations, 94% were either wt or contained mutations conferring resistance to NRTI or PI and were sensitive to lersivirine, with a geometric mean fold change from the control of 1.0-fold (95% CI, 0.9- to 1.2-fold change). Sensitivity to lersivirine was also retained in 62% of viruses containing only 1 NNRTI mutation (21% for efavirenz), 20% of viruses containing 2 NNRTI mutations (4% for efavirenz), and 11% of viruses containing ≥3 NNRTI mutations (0% for efavirenz).
TABLE 3.
No. of NNRTI mutations and lersivirine, efavirenz (EFV), delavirdine (DLV), and nevirapine (NVP) resistance profilesa
| No. of NNRTI mutations (n) | % of viruses remaining sensitive to indicated drug (BCO) |
|||
|---|---|---|---|---|
| Lersivirine (3.1) | EFV (1.75) | NVP (6.63) | DLV (5.86) | |
| 0 (63) | 94 | 89 | 95 | 95 |
| 1 (147) | 62 | 21 | 24 | 40 |
| 2 (90) | 20 | 4 | 2 | 17 |
| ≥3 (53) | 11 | 0 | 0 | 13 |
A total of 353 cloned clinical isolates in Virco's Antivirogram assay (191 viruses) and in Monogram Bioscience's PhenoSense assay (162 viruses) were evaluated. Viruses have been grouped according to the number of NNRTI-specific mutations detected by genotypic analysis. In each case the percentage of viruses that remained sensitive (i.e., had an EC50 <10 times that of the control virus) is shown.
TABLE 4.
Type of HIV-1 isolate mutation and lersivirine, efavirenz (EFV), delavirdine (DLV), and nevirapine (NVP) resistance profilesa
| HIV-1 isolate mutation (n) | % of viruses remaining sensitive to indicated drug (BCO) |
|||
|---|---|---|---|---|
| Lersivirine (3.1) | EFV (1.75) | NVP (6.63) | DLV (5.86) | |
| None (63) | 94 | 89 | 95 | 95 |
| Any (290) | 40 | 12 | 13 | 28 |
| K103N alone (69) | 80 | 7 | 10 | 13 |
| K103N with any mutation but not Y181C or G190A (127) | 48 | 4 | 6 | 7 |
| Y181C alone (14) | 57 | 43 | 14 | 14 |
| Y181C with any mutation but not K103N or G190A (22) | 45 | 32 | 9 | 10 |
| G190A alone (13) | 46 | 0 | 0 | 100 |
| G190A with any mutation but not K103N or Y181C (22) | 36 | 0 | 0 | 91 |
Viruses have been grouped according to the absence of any NNRTI-specific mutation, the presence of any NNRTI-specific mutation, and the presence of one of three mutations, K103N, Y181C, or G190A, either alone or in the presence of any other mutation except the corresponding other two mutations from the K103N, Y181C, and G190A mutations. Mutations K103N, Y181C, and G190A are of key clinical relevance as they occur in the majority of patients failing NNRTI therapy. Table 1 lists the mutations used for this analysis. For each genotype, the percentage of viruses that remained sensitive (i.e., an EC50 <10 times that of HIV NL4-3 control virus) is shown.
Consistent with its binding mode and single directed mutant data described above, lersivirine retained activity against 80% of viruses with genotypes containing K103N as a single NNRTI mutation (versus 7% for efavirenz), 57% of viruses with genotypes containing Y181C as a single NNRTI mutation (43% for efavirenz), and 46% of viruses with genotypes containing G190A as a single NNRTI mutation (0% for efavirenz). This improved resistance profile of lersivirine relative to efavirenz was retained for viruses in which the primary mutations were present with other NNRTI-resistant mutations: for K103N, 48% of viruses were sensitive to lersivirine versus 4% in the case of efavirenz; for Y181C, 45% viruses were sensitive versus 32%; and for G190A, 36% viruses were sensitive versus 0% in the presence of any other NNRTI mutations. Overall, the data from Virco's Antivirogram and Monogram Bioscience's PhenoSense assays demonstrate that lersivirine retains inhibitory activity against a range of clinically derived NNRTI-resistant viruses, in particular viruses containing the substitution K103N, Y181C, or G190A, either alone or in combination with other NNRTI mutations.
Lersivirine is active in primary blood cell assays and against viruses of different clades and diverse geographical origins.
The antiviral activity of lersivirine was assessed in acutely infected PBL. In these experiments, lersivirine inhibited the replication of strain Ba-L in PBL, with a geometric mean EC50 equal to 3.38 nM (95% CI, 2.26 to 5.05 nM) and an EC90 equal to 9.87 nM (95% CI, 6.63 to 14.7 nM), with no cytotoxicity observed up to 50 μM (median CC30 > 96.9 μM, lower quartile of 82.4 μM and upper quartile of >200 μM).
The activity of lersivirine against viruses representing different viral clades was also investigated using a panel of 45 pseudotyped clade B and nonclade B viruses from different geographical origins in the PhenoSense assay (Table 5). All viruses were found to be susceptible to lersivirine, with EC50 fold changes within 2-fold of that for the reference virus, with the exception of one CRF BF and one clade C strain; both demonstrated fold change values within 10-fold (2.3- and 6.3-fold changes, respectively).
TABLE 5.
Lersivirine activity against different HIV-1 cladesa
| HIV-1 clade (n) | Geographical origin(s) | Lersivirine activity |
|
|---|---|---|---|
| EC50 fold change | 95% CI range | ||
| A (A1, AE, AG) (6) | Australia, Canada, Netherlands, Poland, South Africa, United Kingdom | 1.12 | 0.89-1.40 |
| B (5) | Australiad, Canada, Poland, United States | 0.82 | 0.54-1.26 |
| BF (5) | Argentinae | 1.02b | 0.51-2.02 |
| C (25) | South Africaf, Australia, Canada, Puerto Rico, United Kingdom, United States | 1.07c | 0.88-1.30 |
| CH (1) | South Africa | 0.62 | NA |
| D (1) | United Kingdom | 1.02 | NA |
| F1 (1) | Argentina | 1.54 | NA |
| G (1) | United Kingdom | 1.05 | NA |
| Total (45) | 1.03 | 0.91-1.18 | |
Sensitivity of clinically derived virus isolates representing different viral subtypes from treatment-naïve patients to lersivirine. Lersivirine was profiled against 45 cloned clinical isolates in Monogram Bioscience's PhenoSense assay. Geometric mean EC50 fold changes from that for the wt reference virus are reported with 95% confidence interval (CI) ranges. NA, not available.
One circulating recombinant form BF virus showed an EC50 fold change of 2.3.
One clade C virus showed an EC50 fold change of 6.3.
Two clade B viruses tested from Australia.
Five clade BF viruses tested from Argentina.
Twenty clade C viruses tested from South Africa.
Development of in vitro viral resistance to lersivirine requires a distinct resistance pathway.
To characterize the genotypic pathway leading to lersivirine resistance, NL4-3 virus was cultured in SupT1 cells using increasing concentrations of lersivirine (see Fig. S1 in the supplemental material). By week 11, the virus was able to grow in concentrations of lersivirine that were up to 500-fold higher than the original EC50. Two independent virus cultures led to distinct paths to resistance (Table 6), with EC50 shifts of 60.5-fold (“path a”) and 143-fold (“path b”) when tested in the PhenoSense assay. Sequencing of the corresponding RT genes revealed that at passage 9, substitutions V108I and E138K appeared in association with the development of resistance to lersivirine; however, resistance to lersivirine may have occurred at an earlier passage. By passage 11 (path a), the level of resistance had increased further with the addition of F227F/L, while virus from path b contains V108I in association with three further RT mutations: H221Y, F227F/L, and M230M/I.
TABLE 6.
Lersivirine-resistant virus carrying multiple RT mutations and inhibited by other NNRTIsa
| Virus stockb | Passage no.c | RT mutations | Fold change in EC50 from that for drug-sensitive reference virus of indicated drugd |
||||
|---|---|---|---|---|---|---|---|
| Lersivirine | EFV | DLV | NVP | ZDV | |||
| Passaged control | 11 | 0.6 | 0.3 | 0.3 | 0.2 | 0.2 | |
| Lersivirine+ | 9 (a) | V108I, E138K | 14.0 | 1.0 | 1.5 | 0.9 | 0.4 |
| 11 (a) | V108I, E138K, F227F/L | 143 | 3.2 | 1.8 | 12.2 | 0.6 | |
| 11 (b) | V108I, H221Y, F227F/L, M230 M/I | 60.5 | 2.4 | 0.2 | 6.4 | 0.1 | |
| Efavirenz+ | 11 | L100I, K103Q, H221H/Y | 5.0 | 16.3 | 16.4 | 1.0 | 0.1 |
Mutations were identified in the GeneSeq HIV (PR/RT) assay at Monogram Biosciences. Susceptibilities to lersivirine, efavirenz (EFV), delavirdine (DLV), and nevirapine (NVP) were determined in Monogram Bioscience's PhenoSense assay. The NRTI zidovudine (ZDV) was used as a control. No time points before passage 9 were investigated.
Passaged control, virus passaged in the absence of compound; lersivirine+, virus passaged in the presence of increasing concentrations of lersivirine; efavirenz+, virus passaged in the presence of increasing concentrations of efavirenz.
The two different pathways to resistance are labeled a and b (in parentheses).
The drug-sensitive virus CNDO was used as the reference virus for estimation of fold change in EC50.
To understand the potential genetic pathway to resistance from viruses with preexisting NNRTI resistance-associated mutations, K103N and Y181C mutant NL4-3 viruses (along with a wt control) were cultured in SupT1 cells for up to 21 days using fixed concentrations of lersivirine or etravirine. Consistent with the data presented above, the V108I substitution was selected in the presence of lersivirine and was associated with an approximately 5-fold-reduced susceptibility, whether on a wt or K103N RT background (Table 7). Phenotypically, the V108I mutant did not show cross-resistance to etravirine or efavirenz, regardless of the background on which it was selected. In viruses with a Y181C background, a V179V/D mutation in the RT gene was selected both with and without V108I. These viruses show an 8-fold (for the triple mutant) and 11-fold (for the double mutant) decrease in susceptibility to lersivirine. The V179V/D mutation showed between a 7- and 13-fold decrease in susceptibility to both etravirine and efavirenz, respectively. Culture of the K103N virus in the presence of etravirine selected a different set of substitutions in RT (L100I or M230M/I) (data not shown), suggesting a unique genotypic resistance pattern for lersivirine compared with that for either etravirine or efavirenz.
TABLE 7.
Susceptibilities of lersivirine-resistant HIV-1 isolates to lersivirine, efavirenz, and etravirinea
| Virus backboneb | Experimental condition | RT mutation(s) | Fold change in EC50 from that for wt virus of indicated drugc |
||
|---|---|---|---|---|---|
| Lersivirine | EFV | ETV | |||
| wt | Virus after passage through lersivirine | V108I | 5.5 | 1.5 | 0.9 |
| K103N | Input virus | K103N | 2.7 | 35.5 | 1.4 |
| Virus after passage through lersivirine | K103N, V108V/I | 5.0 | 54.8 | 1.1 | |
| Y181C | Input virus | Y181C | 1.8 | 1.9 | 4.4 |
| Virus after passage through lersivirine | Y181C, V179V/D | 11.1 | 9.5 | 13.0 | |
| Virus after passage through lersivirine | Y181C, V108V/I, V179V/D | 8.0 | 7.2 | 9.4 | |
Mutations were identified in the GeneSeq HIV assay at Monogram Biosciences. Susceptibilities to lersivirine, efavirenz (EFV), and etravirine (ETV) were determined in Monogram Biosciences's PhenoSense assay.
Serial passage of wt virus or K103N or Y181C site-directed mutants in fixed concentrations of lersivirine. The wt and K103N and Y181C mutant viruses were passaged at 17 days, 10 days, and 17 days, respectively.
The drug-sensitive virus CNDO was used as the reference virus for estimation of fold change in EC50 in the PhenoSense assay; this value was then used to determine the EC50 fold change from that for the wt virus.
Lersivirine is additive or synergistic with other antiretroviral agents.
To establish the potential of lersivirine for administration in combination with other antiretroviral agents, the in vitro antiviral effect of the pair-wise combinations of lersivirine with class representatives of currently approved and investigational anti-HIV agents was assessed using MT-2 cells and measuring cell viability 5 days after NL4-3 infection (Table 8). Combinations of lersivirine with drugs of the NRTI class (abacavir, didanosine, emtricitabine, lamivudine, tenofovir, and zidovudine) resulted in synergistic interactions. In addition, synergistic interactions were observed with the integrase inhibitors elvitegravir and raltegravir and the fusion inhibitor enfuvirtide. Lersivirine in combination with a PI generally resulted in additive interactions (atazanavir, lopinavir, and ritonavir). There was no evidence of antagonistic interactions with any of the compounds investigated, and there was no evidence of synergistic cytotoxicity at the highest concentrations tested (1 μM for lersivirine).
TABLE 8.
Synergy/additivity of lersivirine with agents from other antiretroviral classes in vitroa
| Agent (n) | Class | Mean vol ± SD (μM2%) of lersivirine: |
||
|---|---|---|---|---|
| Synergy | Antagonism | Combined effect | ||
| Abacavir (3) | NRTI | 90.6 ± 26.5 | −49.3 ± 84.2 | Moderate synergy (minor antagonism) |
| Didanosine (4) | NRTI | 156.0 ± 66.1 | −3.7 ± 7.3 | Strong synergy |
| Emtricitabine (4) | NRTI | 140.5 ± 57.1 | −5.1 ± 8.9 | Strong synergy |
| Lamivudine (4) | NRTI | 175.1 ± 81.1 | −6.3 ± 12.1 | Strong synergy |
| Tenofovir (3) | NRTI | 115.6 ± 40.7 | −10.8 ± 11.9 | Strong synergy |
| Zidovudine (3) | NRTI | 177.5 ± 183.8 | −15.4 ± 26.7 | Strong synergyb |
| Atazanavir (2) | PI | 6.6 ± 9.4 | −23.1 ± 32.6 | Additive |
| Lopinavir (2) | PI | 29.1 ± 19.4 | −28.5 ± 30.3 | Minor synergy/minor antagonismc |
| Ritonavir (2) | PI | 12.6 ± 0.6 | −19.1 ± 27.1 | Additive |
| Elvitegravir (3) | INT | 69.7 ± 66.5 | −20.1 ± 34.8 | Moderate synergy |
| Raltegravir (2) | INT | 34.6 ± 13.8 | −10.7 ± 15.1 | Minor synergy |
| Enfuvirtide (3) | Entry | 75.1 ± 77.8 | −32.8 ± 56.8 | Moderate synergy (minor antagonism) |
Experiments performed using a cytopathic-effect-based assay with HIV-1 NL4-3 in MT-2 cells. Volumes of synergy (μM2%) were calculated at 95% confidence intervals using drug combination data from three to four replicate plates per assay, with the aid of MacSynergy II software. Volumes are expressed as means from two to four independent experiments (± standard deviations [SD]). For these studies, synergy (assigned a positive value) or antagonism (assigned a negative value) was defined as drug combinations yielding mean volumes in excess of 25 μM2%. Moderate synergistic/antagonistic activity and strong synergistic/antagonistic activity were defined as mean volumes between 50 and 100 μM2% and in excess of 100 μM2%, respectively. Additive drug interactions were defined by mean volumes of 0 to 25 μM2%. Elvitegravir is an investigational integrase inhibitor (INT).
Strong synergy seen in two experiments; additive interaction in the third experiment.
Minor synergy/minor antagonism seen in one experiment; additive interaction in the second experiment.
DISCUSSION
Since the first agent, zidovudine, was licensed for the treatment of HIV-1 infection in 1987, a total of 25 drugs have been approved for the treatment of HIV (13). Since 1996, the importance of anti-HIV drug combination regimens has become widely accepted, and in 2008, the Department of Health and Human Services guidelines (46) recommend the inclusion of at least two, and preferably three, fully active antiretroviral agents when constructing drug regimens. Efavirenz-based therapy or boosted PI-based therapies are currently recommended as first-line regimens for the treatment of HIV infection. Although these recommendations provide effective treatment options for the majority of patients, treatment durability is still limited by drug-related side effects, inadequate patient adherence, and the development of drug resistance.
NNRTI-based regimens have been associated with a low genetic barrier to resistance, rash, hypersensitivity reactions, and central nervous system side effects. Etravirine, a novel NNRTI with activity against a broader panel of NNRTI mutant viruses, has recently been approved for use in treatment-experienced patients who have evidence of infection with HIV-1 strains that are resistant to an NNRTI and other antiretroviral agents (Intelence prescribing information; Tibotec Therapeutics, Raritan, NJ). However, rash and nausea are commonly reported adverse events in etravirine-treated subjects. There is, therefore, still a need for first-line antiviral agents that will facilitate patient adherence and allow durable suppression of viral replication.
In our laboratories, we have recently identified a novel inhibitor of HIV (11). In this paper, we confirm that lersivirine belongs to the NNRTI class: in vitro, it inhibits the RT enzyme by a mixed noncompetitive kinetic mechanism and binds the protein with a 1:1 stoichiometry in the NNRTI binding pocket. X-ray crystallography suggests a novel binding mode of lersivirine into the “NNRTI pocket,” which in turn translates to activity against key class resistance mutants.
A survey of more than 5,000 patient samples of NNRTI-treated individuals revealed that the most common mutations were K103N (29%), Y181C (14%), and G190A (17%) (6). In agreement with this distribution, the most commonly occurring NNRTI mutations found in our panels of 353 isolates tested were K103N > Y181C/I > G190A/S. In this panel, lersivirine was active against all clinically derived viruses that had no NNRTI mutations, with many having phenotypic resistance to antiviral drugs of the NRTI and PI classes (data not shown).
Furthermore, lersivirine also retained inhibitory activity against a range of viruses carrying clinically relevant NNRTI mutations. Lersivirine inhibited 62% of isolates containing a single NNRTI mutation (in vitro resistance was defined using the BCO of 3.1) and was active against the majority of viruses carrying the single mutations K103N, Y181C, and G190A, which occur in the majority of patients failing NNRTI therapy (Table 6). Although clinical studies have yet to confirm this finding in patients, these initial results suggest that lersivirine has the potential to be used in patients that failed a drug regimen containing efavirenz, nevirapine, or delavirdine because of mutations arising at location K103, Y181, or G190.
In our subtype study, one clade C virus (isolated in Puerto Rico) demonstrated a 6.3-fold IC50 to the reference virus strain. Population sequence analysis did not reveal the presence of the V106M mutation, which is often associated with resistance to efavirenz and NVP in clade C viruses, or of any other NNRTI resistance-associated mutations (21). However, a number of amino acid changes were present in the RT gene from this patient virus, which may result in differences in susceptibility but not necessarily resistance. These include amino acids at positions 35, 135, 158, and 245 (9; J. Whitcomb, personal communication). Slight decreases in susceptibility were also observed when this isolate was tested against efavirenz and nevirapine (1.9- and 2.2-fold changes from those for the reference virus, respectively).
Sequential passage experiments with wt virus in the presence of progressively increasing concentrations of drug in vitro can identify the accumulation of mutations that confer resistance. Lersivirine dose escalation studies identified two pathways to resistance; in both cases V108I and F227L were selected, with resistance increased further as a result of the emergence of E138K or M230M/I. The V108I mutation was also selected in fixed-dose studies using the wt and viruses with preexisting NNRTI-resistant mutations. Acquisition of a V108I mutation appears to be the preferred molecular pathway to lersivirine resistance and differs from that of efavirenz, etravirine, and rilpivirine. Two different pathways to resistance were reported following dose escalation selection experiments with WT virus for etravirine: V179F plus Y181C plus M230L and Y181C plus G190E (49). In addition, the Y181C mutation was selected in etravirine fixed-dose studies, suggesting the importance of this mutation in the development of resistance to etravirine (49).
Several pathways were identified following escalating dose concentrations of rilpivirine; combinations of the mutations L100I, K101E, Y181C, and F227V were frequently seen with the laboratory strain IIIB (4). The preferred molecular pathway to lersivirine resistance would appear to differ from that of etravirine and rilpivirine. The most common NNRTI resistance-associated substitutions (seen in at least 10% of patients) that emerged in patients with virologic failure—who received etravirine in the DUET-1 and -2 studies—included V108I, V179F, V179I, and Y181C. These usually emerged in a background of other NNRTI resistance-associated substitutions (20, 43).
Interestingly, phenotypic data indicated that multiple mutations were required to establish substantial resistance to lersivirine. In both dose escalation and fixed-dose studies V108I was selected, suggesting this variant to be the preferred molecular pathway under lersivirine drug pressure. Phenotypically, the V108I mutant showed no cross-resistance to efavirenz or etravirine. Similarly, lersivirine-resistant viruses generated by dose escalation showed no cross-resistance to efavirenz and DLV. These results suggest that resistance was associated with, and specific for, the presence of lersivirine and may not lead to class-wide resistance.
The question arises as to what makes lersivirine resilient against drug resistance mutations. While it is difficult to unambiguously rationalize the effect of mutations in the NNRTI binding pocket with changes in IC50, the three-dimensional structure that we describe allows us to propose a hypothesis. As shown in Fig. 3C, lersivirine makes extensive contacts through the 3,5-dicyanophenoxy group with Trp229 at the top of the binding pocket. This residue not only is part of the NNRTI binding pocket but also belongs to the “primer grip” of HIV RT, and any mutation at this position has been shown to severely compromise the RNA- and DNA-dependent DNA polymerase activities of the enzyme (33). Indeed, this immutable residue has become the focus of the development of novel NNRTIs (17). In addition, upon the binding of many NNRTIs, the side chain of Tyr181, contained within the β4-β7-β8 sheet, have been reported to flip conformation from a “down” to an “up” position (38). Upon the binding of lersivirine into the NNRTI binding pocket, the Tyr181 is in the less frequently observed “down” conformation. This tyrosine does not make the equivalent contacts with the disubstituted group that is observed with other NNRTIs, such as capravirine (37). The resilience of lersivirine to the Y181C mutation can be explained by a reduction in the aromatic stacking interactions with these residues. A recently reported benzophenone NNRTI series also takes advantage of this alternative Y181 conformation (35).
Finally, we also observed that specific polar interactions between RT and lersivirine are limited to the hydrogen bonds formed to the backbone amides of the protein. We found that lersivirine accepts a hydrogen bond from the Lys103 amide and donates a hydrogen bond to the Pro236 carbonyl. No specific interaction is made with the 103 side chain, and no significant change in compound binding to the mutant enzyme is observed. In short, lersivirine is an expression of a minimal NNRTI pharmacophore in which maximum advantage is taken of the regions of the binding sites that are constant while minimizing contacts with regions of the site that are known to mutate. This strategy differs from that adopted for higher-molecular-weight NNRTIs, such as etravirine, where mutations are compensated for by a larger contact area. The susceptibility of lersivirine to the V108I mutant (a 5-fold decrease in potency) can be rationalized as this residue makes extensive van der Waals contact with the dicyano-substituted phenol ring (4.0-Å contact distance to the ring). A sterically more demanding isoleucine at this position could perturb the binding mode observed.
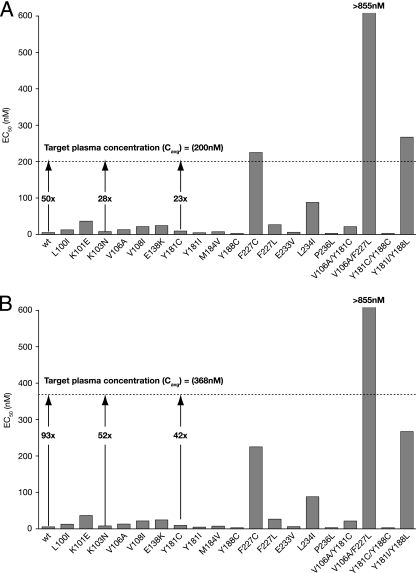
Taking these data into consideration, lersivirine should be a potent agent active against key clinical mutant HIV-1 strains. Indeed, when the EC50 for lersivirine is compared to the steady-state pharmacokinetics of lersivirine in HIV-1-infected patients receiving the compound as monotherapy at 500 mg once a day (QD) or 750 mg QD, there is a 50- and 93-fold window, respectively, between the protein-adjusted wt geometric mean inhibition in vitro and the average free drug levels achieved (Fig. 5) (15, 16). Interestingly, at these doses, the average free concentrations of lersivirine would be expected to be well above the EC50 for most mutants studied.
FIG. 5.
Comparison of the free steady-state average concentrations (Cavg) in the monotherapies with 500 mg QD (A) and 750 mg QD (B) in HIV patients and activity of lersivirine (EC50 potency) against wt and mutant NL4-3 viruses (Fig. 4B). Target-free plasma Cavg is calculated using the area under the concentration-time curve from 0 to 24 h (AUC0-24) on day 8 [AUCtau = 3,543.5 ng·h/ml for the 500 mg QD (147.6 ng/ml; n = 6) (A) and AUCtau = 6,530.1 ng·h/ml for the 750 mg QD (272.1 ng/ml; n = 6) (B)] and takes into account plasma protein binding (∼60%) and the molecular weight of lersivirine (310.4). The fold changes (Cavg/EC50) for the wt and K103N and Y181C mutants are shown by arrows.
To support the use of lersivirine as part of a multidrug regimen, the antiviral activity of lersivirine was evaluated in combination with representatives from each of the licensed antiretroviral classes in vitro. Preliminary data indicate additive antiviral interactions with all compounds in combination with lersivirine (with the exception of the NRTIs, which frequently demonstrated synergistic interactions). Synergy has previously been reported between NRTIs and NNRTIs which target the same viral enzyme. Zidovudine and efavirenz were shown to be synergistic in cell culture, and studies of etravirine combined with zidovudine suggested synergy, though additive effects were reported when etravirine was combined with other NRTI drugs (abacavir, didanosine, lamivudine, stavudine, zalcitabine, zidovudine, and tenofovir) and representatives of the PIs and the currently marketed NNRTIs (2, 26). The synergistic interaction between NRTIs and NNRTIs has been associated with the ability of the NNRTI to inhibit ATP-mediated removal of the NRTI and so prolong its MOA (7). The synergistic behavior of NRTIs and NNRTIs in combination in vitro may contribute to their effectiveness in vivo.
There was no evidence of antagonistic interactions with any of the compounds investigated, and there was no evidence of synergistic cytotoxicity at the highest concentrations tested.
In conclusion, lersivirine represents an attractive novel NNRTI with potency against an interesting panel of key clinical NNRTI mutant HIV-1 viruses. Ongoing clinical trials (15) are investigating its potential for treatment of HIV-1-infected patients in the context of HAART.
Supplementary Material
Acknowledgments
R. Corbau, J. Mori, C. Phillips, L. Fishburn, A. Martin, C. Mowbray, W. Panton, C. Smith-Burchnell, A. Thornberry, H. Ringrose, T. Knöchel, S. Irving, M. Westby, A. Wood, and M. Perros are current or former employees of Pfizer Global Research and Development.
The support provided by Iain Nicholson at Complete Medical Communications funded by Pfizer Inc. consisted solely of manuscript editing and formatting, and no contribution was made to editorial content.
Footnotes
Published ahead of print on 26 July 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andries, K., H. Azijn, T. Thielemans, D. Ludovici, M. Kukla, J. Heeres, P. Janssen, B. De Corte, J. Vingerhoets, R. Pauwels, and M. P. de Bethune. 2004. TMC125, a novel next-generation nonnucleoside reverse transcriptase inhibitor active against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:4680-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antinori, A., M. Zaccarelli, A. Cingolani, F. Forbici, M. G. Rizzo, M. P. Trotta, S. Di Giambenedetto, P. Narciso, A. Ammassari, E. Girardi, A. De Luca, and C. F. Perno. 2002. Cross-resistance among nonnucleoside reverse transcriptase inhibitors limits recycling efavirenz after nevirapine failure. AIDS Res. Hum. Retroviruses 18:835-838. [DOI] [PubMed] [Google Scholar]
- 4.Azijn, H., I. Tirry, J. Vingerhoets, M. P. de Bethune, G. Kraus, K. Boven, D. Jochmans, E. Van Craenenbroeck, G. Picchio, and L. T. Rimsky. 2010. TMC278, a next-generation nonnucleoside reverse transcriptase inhibitor (NNRTI), active against wild-type and NNRTI-resistant HIV-1. Antimicrob. Agents Chemother. 54:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey, S. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 6.Balzarini, J. 2004. Current status of the non-nucleoside reverse transcriptase inhibitors of human immunodeficiency virus type 1. Curr. Top. Med. Chem. 4:921-944. [DOI] [PubMed] [Google Scholar]
- 7.Basavapathruni, A., J. Vingerhoets, M. P. de Bethune, R. Chung, C. M. Bailey, J. Kim, and K. S. Anderson. 2006. Modulation of human immunodeficiency virus type 1 synergistic inhibition by reverse transcriptase mutations. Biochemistry 45:7334-7340. [DOI] [PubMed] [Google Scholar]
- 8.Best, B. M., and M. Goicoechea. 2008. Efavirenz—still first-line king? Expert Opin. Drug Metab. Toxicol. 4:965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, A. J., H. M. Precious, J. M. Whitcomb, J. K. Wong, M. Quigg, W. Huang, E. S. Daar, R. T. D'Aquila, P. H. Keiser, E. Connick, N. S. Hellmann, C. J. Petropoulos, D. D. Richman, and S. J. Little. 2000. Reduced susceptibility of human immunodeficiency virus type 1 (HIV-1) from patients with primary HIV infection to nonnucleoside reverse transcriptase inhibitors is associated with variation at novel amino acid sites. J. Virol. 74:10269-10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buckheit, R. W., Jr., K. Watson, V. Fliakas-Boltz, J. Russell, T. L. Loftus, M. C. Osterling, J. A. Turpin, L. A. Pallansch, E. L. White, J. W. Lee, S. H. Lee, J. W. Oh, H. S. Kwon, S. G. Chung, and E. H. Cho. 2001. SJ-3366, a unique and highly potent nonnucleoside reverse transcriptase inhibitor of human immunodeficiency virus type 1 (HIV-1) that also inhibits HIV-2. Antimicrob. Agents Chemother. 45:393-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burt, C., R. Corbau, S. Gayton, M. Hawes, L. H. Jones, C. E. Mowbray, M. Perros, I. Tran, D. Price, F. Quinton, M. Selby, P. Stupple, R. Webster, and A. Wood. 2009. Pyrazole NNRTIs 4: selection of UK-453,061 (Lersivirine) as a development candidate. Bioorg. Med. Chem. Lett. 19:5857-5860. [DOI] [PubMed] [Google Scholar]
- 12.Corbett, J. W., S. S. Ko, J. D. Rodgers, S. Jeffrey, L. T. Bacheler, R. M. Klabe, S. Diamond, C. M. Lai, S. R. Rabel, J. A. Saye, S. P. Adams, G. L. Trainor, P. S. Anderson, and S. K. Erickson-Viitanen. 1999. Expanded-spectrum nonnucleoside reverse transcriptase inhibitors inhibit clinically relevant mutant variants of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 43:2893-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Clercq, E. 2009. Anti-HIV drugs: 25 compounds approved within 25 years after the discovery of HIV. Int. J. Antimicrob. Agents 33:307-320. [DOI] [PubMed] [Google Scholar]
- 14.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 15.Fätkenheuer, G., S. Staszewski, A. Plettenburg, F. Hackman, G. Layton, L. McFadyen, J. Davis, and T. M. Jenkins. 2009. Activity, pharmacokinetics and safety of lersivirine (UK-453,061), a next-generation nonnucleoside reverse transcriptase inhibitor, during 7-day monotherapy in HIV-1-infected patients. AIDS 23:2115-2122. [DOI] [PubMed] [Google Scholar]
- 16.Fätkenheuer, G., S. Staszewski, A. Plettenburg, F. Hackman, G. Layton, L. McFadyen, J. Davis, and T. Jenkins. 2007. Short-term monotherapy with UK-453,061, a novel NNRTI, reduces viral load in HIV-infected patients. Proceedings of the 4th International AIDS Society Conference on HIV Pathogenesis, Treatment and Prevention, Chicago, IL.
- 17.Fattorusso, C., S. Gemma, S. Butini, P. Huleatt, B. Catalanotti, M. Persico, M. De Angelis, I. Fiorini, V. Nacci, A. Ramunno, M. Rodriquez, G. Greco, E. Novellino, A. Bergamini, S. Marini, M. Coletta, G. Maga, S. Spadari, and G. Campiani. 2005. Specific targeting highly conserved residues in the HIV-1 reverse transcriptase primer grip region. Design, synthesis, and biological evaluation of novel, potent, and broad spectrum NNRTIs with antiviral activity. J. Med. Chem. 48:7153-7165. [DOI] [PubMed] [Google Scholar]
- 18.Freimuth, W. W. 1996. Delavirdine mesylate, a potent non-nucleoside HIV-1 reverse transcriptase inhibitor. Adv. Exp. Med. Biol. 394:279-289. [DOI] [PubMed] [Google Scholar]
- 19.Gao, H. Q., P. L. Boyer, E. Arnold, and S. H. Hughes. 1998. Effects of mutations in the polymerase domain on the polymerase, RNase H and strand transfer activities of human immunodeficiency virus type 1 reverse transcriptase. J. Mol. Biol. 277:559-572. [DOI] [PubMed] [Google Scholar]
- 20.Geretti, A. M. 2008. Shifting paradigms: the resistance profile of etravirine. J. Antimicrob. Chemother. 62:643-647. [DOI] [PubMed] [Google Scholar]
- 21.Hall, D. B., F. van Leth, P. Robinson, P. McKenna, F. W. M. N. Wit, and J. M. A. Lange. 2004. The V106M mutation in treatment failures from a randomized controlled trial of lamivudine and stavudine with nevirapine and/or efavirenz, p. S177. In Antiviral therapy. Proceedings of the XIIIth International HIV Drug Resistance Workshop: Basic Principles & Clinical Implications, Tenerife Sur-Costa Adeje, Canary Islands, Spain.
- 22.Harris, M. 2003. Efficacy and durability of nevirapine in antiretroviral-experienced patients. J. Acquir Immune Defic Syndr 34(Suppl. 1):S53-S58. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, V. A., F. Brun-Vezinet, B. Clotet, H. F. Gunthard, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2008. Update of the Drug Resistance Mutations in HIV-1. Top. HIV Med. 16:138-145. [PubMed] [Google Scholar]
- 24.Jones, L. H., G. Allan, O. Barba, C. Burt, R. Corbau, T. Dupont, T. Knochel, S. Irving, D. S. Middleton, C. E. Mowbray, M. Perros, H. Ringrose, N. A. Swain, R. Webster, M. Westby, and C. Phillips. 2009. Novel indazole non-nucleoside reverse transcriptase inhibitors using molecular hybridization based on crystallographic overlays. J. Med. Chem. 52:1219-1223. [DOI] [PubMed] [Google Scholar]
- 25.Kim, B., T. R. Hathaway, and L. A. Loeb. 1998. Fidelity of mutant HIV-1 reverse transcriptases: interaction with the single-stranded template influences the accuracy of DNA synthesis. Biochemistry 37:5831-5839. [DOI] [PubMed] [Google Scholar]
- 26.King, R. W., R. M. Klabe, C. D. Reid, and S. K. Erickson-Viitanen. 2002. Potency of nonnucleoside reverse transcriptase inhibitors (NNRTIs) used in combination with other human immunodeficiency virus NNRTIs, NRTIs, or protease inhibitors. Antimicrob. Agents Chemother. 46:1640-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knoll, B. M., S. Vento, and Z. Temesgen. 2008. Etravirine. Drugs Today (Barcelona) 44:23-33. [DOI] [PubMed] [Google Scholar]
- 28.Lowe, D. M., A. Aitken, C. Bradley, G. K. Darby, B. A. Larder, K. L. Powell, D. J. Purifoy, M. Tisdale, and D. K. Stammers. 1988. HIV-1 reverse transcriptase: crystallization and analysis of domain structure by limited proteolysis. Biochemistry 27:8884-8889. [DOI] [PubMed] [Google Scholar]
- 29.Maggiolo, F. 2007. Efavirenz. Expert Opin. Pharmacother. 8:1137-1145. [DOI] [PubMed] [Google Scholar]
- 30.Mellors, J., and S. Kemp. 2001. Experts discuss findings on drug resistance. AIDS Alert 16:103-104. [PubMed] [Google Scholar]
- 31.Milinkovic, A., and E. Martinez. 2004. Nevirapine in the treatment of HIV. Expert Rev. Anti. Infect. Ther. 2:367-373. [DOI] [PubMed] [Google Scholar]
- 32.Miller, V., M. P. de Bethune, A. Kober, M. Sturmer, K. Hertogs, R. Pauwels, P. Stoffels, and S. Staszewski. 1998. Patterns of resistance and cross-resistance to human immunodeficiency virus type 1 reverse transcriptase inhibitors in patients treated with the nonnucleoside reverse transcriptase inhibitor loviride. Antimicrob. Agents Chemother. 42:3123-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelemans, H., R. Esnouf, E. De Clercq, and J. Balzarini. 2000. Mutational analysis of trp-229 of human immunodeficiency virus type 1 reverse transcriptase (RT) identifies this amino acid residue as a prime target for the rational design of new non-nucleoside RT inhibitors. Mol. Pharmacol. 57:954-960. [PubMed] [Google Scholar]
- 34.Prichard, M. N., and C. Shipman, Jr. 1990. A three-dimensional model to analyze drug-drug interactions. Antiviral Res. 14:181-205. [DOI] [PubMed] [Google Scholar]
- 35.Ren, J., P. P. Chamberlain, A. Stamp, S. A. Short, K. L. Weaver, K. R. Romines, R. Hazen, A. Freeman, R. G. Ferris, C. W. Andrews, L. Boone, J. H. Chan, and D. K. Stammers. 2008. Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors. J. Med. Chem. 51:5000-5008. [DOI] [PubMed] [Google Scholar]
- 36.Ren, J., J. Milton, K. L. Weaver, S. A. Short, D. I. Stuart, and D. K. Stammers. 2000. Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase. Structure 8:1089-1094. [DOI] [PubMed] [Google Scholar]
- 37.Ren, J., C. Nichols, L. E. Bird, T. Fujiwara, H. Sugimoto, D. I. Stuart, and D. K. Stammers. 2000. Binding of the second generation non-nucleoside inhibitor S-1153 to HIV-1 reverse transcriptase involves extensive main chain hydrogen bonding. J. Biol. Chem. 275:14316-14320. [DOI] [PubMed] [Google Scholar]
- 38.Ren, J., C. E. Nichols, P. P. Chamberlain, K. L. Weaver, S. A. Short, J. H. Chan, J. P. Kleim, and D. K. Stammers. 2007. Relationship of potency and resilience to drug resistant mutations for GW420867X revealed by crystal structures of inhibitor complexes for wild-type, Leu100Ile, Lys101Glu, and Tyr188Cys mutant HIV-1 reverse transcriptases. J. Med. Chem. 50:2301-2309. [DOI] [PubMed] [Google Scholar]
- 39.Roversi, P., E. Blanc, C. Vonrhein, G. Evans, and G. Bricogne. 2000. Modelling prior distributions of atoms for macromolecular refinement and completion. Acta Crystallogr. D Biol. Crystallogr. 56:1316-1323. [DOI] [PubMed] [Google Scholar]
- 40.Scott, L. J., and C. M. Perry. 2000. Delavirdine: a review of its use in HIV infection. Drugs 60:1411-1444. [DOI] [PubMed] [Google Scholar]
- 41.Soriano, V., and C. de Mendoza. 2002. Genetic mechanisms of resistance to NRTI and NNRTI. HIV Clin. Trials 3:237-248. [DOI] [PubMed] [Google Scholar]
- 42.Tambuyzer, L., H. Azijn, L. T. Rimsky, J. Vingerhoets, P. Lecocq, G. Kraus, G. Picchio, and M. P. de Bethune. 2009. Compilation and prevalence of mutations associated with resistance to non-nucleoside reverse transcriptase inhibitors. Antivir. Ther. 14:103-109. [PubMed] [Google Scholar]
- 43.Tambuyzer, L., J. Vingerhoets, H. Azijn, B. Daems, G. Picchio, and M. P. de Bethune. 2008. Emerging mutations in patients with virological failure on etravirine (ETR; TMC125) in the DUET-1 and DUET-2 clinical trials. 6th Eur. HIV Drug Resist. Workshop, Budapest, Hungary, 26 to 28 March 2008.
- 44.Tibotec Therapeutics. 2009. Etravirine prescribing information. Tibotec Therapeutics, Raritan, NJ. http://www.accessdata.fda.gov/drugsatfda_docs/label/2009/022187s002lbl.pdf.
- 45.UNAIDS. 2007. 2007 UNAIDS annual report: knowing your epidemic. Joint United Nations Programme on HIV/AIDS, Geneva, Switzerland. http://data.unaids.org/pub/Report/2008/jc1535_annual_report07_en.pdf.
- 46.U.S. Department of Health and Human Services. 3 November 2008. DHHS guidelines. Panel on antiretroviral guidelines for adults and adolescents. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents, p. 1-139. U.S. Department of Health and Human Services, Washington, DC. http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf. Accessed 13 July 2009.
- 47.Verbiest, W. 2001. Poster. 2nd Int. Antiviral Drug Disc. Dev. Summit, Princeton, NJ, 28 to 29 March 2001.
- 48.Verlinden, T., H. Vermeiren, P. Lecocq, L. Bacheler, P. McKenna, M. Vanpachtenbeke, L. I. Laenen-Horvat, M. Van Houtte, and L. J. Stuyver. 2005. Assessment of the Antivirogram performance over time including a revised definition of biological test cut-off values, abstr. 46. Abstr. XIVth Int. HIV Drug Resist. Workshop, Quebec City, Quebec, Canada, 7 to 11 June 2005.
- 49.Vingerhoets, J., H. Azijn, E. Fransen, I. De Baere, L. Smeulders, D. Jochmans, K. Andries, R. Pauwels, and M. P. de Bethune. 2005. TMC125 displays a high genetic barrier to the development of resistance: evidence from in vitro selection experiments. J. Virol. 79:12773-12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young, S. D., S. F. Britcher, L. O. Tran, L. S. Payne, W. C. Lumma, T. A. Lyle, J. R. Huff, P. S. Anderson, D. B. Olsen, and S. S. Carroll. 1995. L-743, 726 (DMP-266): a novel, highly potent nonnucleoside inhibitor of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 39:2602-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.