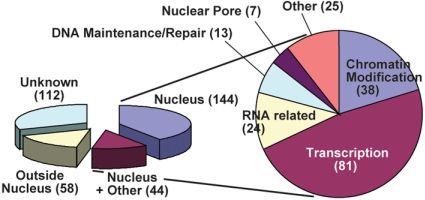
Table 2.
| Functional group | SUMO substrate |
| SUMO pathway | SUMO1, SAE2, SCE1, SIZ1, ESD4 |
| Transcription factors | |
| WRKY family | WRKY3, WRKY4, WRKY6, WRKY33, WRKY72 |
| HSF family | HSFA1D, HSFA2, HSFB2B |
| Global TF family | GT2, GTB1, GTE1, GTE4, GTE7, GTL1 |
| ANACONDA family | ANACO50, ANACO51, ANACO80/ATAF2 |
| Homeodomain | BEL1, BEL10, KNAT3, HB6, ANL2 |
| Others of note | EIN3, ARR1, PHR1, ERF6, BIM1, HUA2 |
| Transciptional coregulators | |
| TOPLESS family | TPL, TPR1, TPR2, TPR4 |
| Others of note | LEUNIG, LUH, SEUSS |
| Chromatin modifiers | |
| Chromatin methylation-related | SUVR1, SUVR2, SUVH2, SUVH9, KTF1, IBM1 |
| SWI/SNF complex | SWI3A, SWI3C, SWI3D, PICKLE, PKR1, CHR11 |
| Histone acetylation-related | SNL2, SNL4, SNL5, HAC1, GCN5, HDA19(HD1), ADA2A, ADA2B |
| Histone 2b-related | Histone2B, NRP1, UBP26, SPT16 |
| DNA main/repair | LIG1, DRT111, KU80, POLD3, TOP1, TRB1 |
| RNA-related | SERRATE, LACHESIS, XRN3, DRH1, SDN3 PRP40B, LA1, PARL1, STA1 |
| Nuclear pore | IMP-6, IMPA1, WIP1 |
| Cell cycle regulators | SYN4, ILP1, ILP2, RHL1, DPB, CDC5, CDC48, RPA1 |
*See Table S2 or the Scaffold file (Dataset S1) available at http://www.genetics.wisc.edu/node/558 for complete list.
†Number of unique proteins are in parentheses.