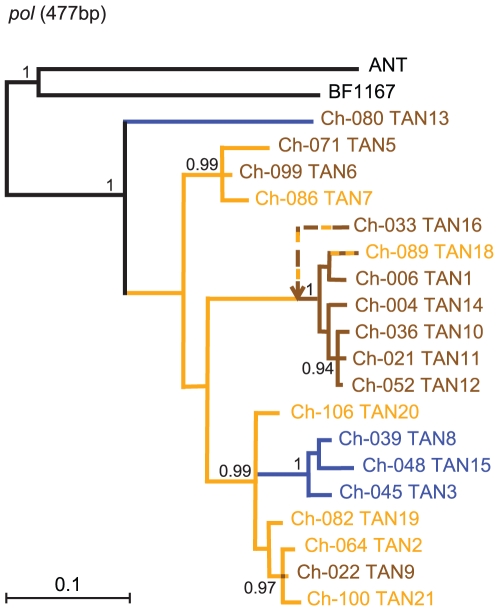
Figure 4. Phylogeny of SIVcpz in Gombe.
A phylogenetic tree was constructed from available pol sequences, using SIVcpzPts strains from the Democratic Republic of Congo (ANT and BF1167) as outgroups. Viruses (and their chimpanzee hosts) are color coded according to their most recent community (Mitumba, blue; Kasekela, brown; Kalande orange), while branch colors indicate the likely origin of these infections (e.g., Ch-099 resided in Kasekela from 2004 to 2006, but acquired TAN6 in Kalande). Striped lines indicate uncertain origin (genetic and demographic data suggest that Ch-033 and Ch-089 acquired their infections in Kalande and Kasekela, respectively; the location where Ch-022 acquired SIVcpz is unknown). The phylogenetic position of TAN16 is approximated (arrow) based on the position of its env-nef sequences (see Figure S2B in reference [2]). The tree was inferred by Bayesian methods [38]; numbers on nodes indicate posterior probabilities (only values above 0.95 are shown). The scale bar represents 0.1 substitutions per site.